Project
DANIO-CODE
Navigation
Downloads
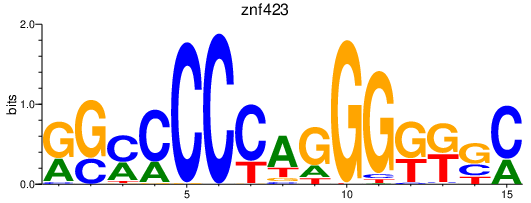
Results for znf423
Z-value: 0.69
Transcription factors associated with znf423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf423
|
ENSDARG00000095732 | zinc finger protein 423 |
Activity profile of znf423 motif
Sorted Z-values of znf423 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of znf423
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_32752138 | 2.94 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr12_+_30245620 | 1.90 |
ENSDART00000153116
ENSDART00000152900 |
ENSDARG00000020224
|
ENSDARG00000020224 |
| chr8_-_52952226 | 1.81 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr21_-_23271127 | 1.63 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr5_+_32231368 | 1.51 |
ENSDART00000169358
|
ENSDARG00000095142
|
ENSDARG00000095142 |
| chr13_-_29875809 | 1.50 |
ENSDART00000049624
|
wnt8b
|
wingless-type MMTV integration site family, member 8b |
| chr8_-_52952141 | 1.24 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr1_-_24805276 | 1.05 |
ENSDART00000102498
|
sfrp2
|
secreted frizzled-related protein 2 |
| chr7_-_26153788 | 0.98 |
|
|
|
| chr5_+_32231323 | 0.93 |
ENSDART00000169358
|
ENSDARG00000095142
|
ENSDARG00000095142 |
| chr12_-_41124276 | 0.65 |
ENSDART00000172022
ENSDART00000158605 ENSDART00000162967 |
dpysl4
|
dihydropyrimidinase-like 4 |
| chr1_+_11485480 | 0.60 |
ENSDART00000132560
|
stra6l
|
STRA6-like |
| chr16_-_31868810 | 0.55 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr2_+_15459785 | 0.55 |
ENSDART00000016071
ENSDART00000141921 |
arhgap29b
|
Rho GTPase activating protein 29b |
| chr25_-_727490 | 0.53 |
|
|
|
| chr10_+_9575 | 0.53 |
|
|
|
| chr7_+_54336324 | 0.51 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr22_+_39067567 | 0.49 |
|
|
|
| chr24_-_26594117 | 0.43 |
ENSDART00000124580
|
pld1b
|
phospholipase D1b |
| chr24_-_26594053 | 0.43 |
ENSDART00000124580
|
pld1b
|
phospholipase D1b |
| chr5_+_61199427 | 0.38 |
ENSDART00000132452
|
sympk
|
symplekin |
| chr14_-_16118606 | 0.36 |
|
|
|
| chr5_-_56856570 | 0.35 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr22_-_6469430 | 0.27 |
ENSDART00000142235
|
si:dkey-19a16.7
|
si:dkey-19a16.7 |
| chr20_-_22293402 | 0.26 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr8_-_47156995 | 0.23 |
|
|
|
| chr8_-_47156727 | 0.23 |
|
|
|
| chr23_+_4423579 | 0.21 |
ENSDART00000039829
|
zgc:112175
|
zgc:112175 |
| chr3_-_57709184 | 0.15 |
ENSDART00000074285
ENSDART00000170918 ENSDART00000171040 |
syngr2a
|
synaptogyrin 2a |
| chr22_+_25534262 | 0.12 |
ENSDART00000131218
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr7_+_54336245 | 0.11 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr4_-_72183874 | 0.03 |
ENSDART00000144382
|
si:dkey-262g12.12
|
si:dkey-262g12.12 |
| chr1_-_53465275 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.4 | 1.6 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.9 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.6 | GO:0021572 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.1 | 0.6 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 3.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 1.1 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 3.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |