Project
DANIO-CODE
Navigation
Downloads
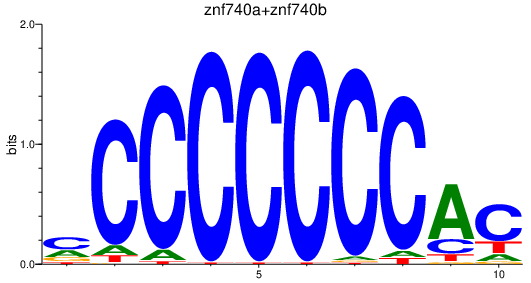
Results for znf740a+znf740b
Z-value: 1.73
Transcription factors associated with znf740a+znf740b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf740b
|
ENSDARG00000061174 | zinc finger protein 740b |
|
znf740a
|
ENSDARG00000070939 | zinc finger protein 740a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf740a | dr10_dc_chr11_-_2274344_2274483 | -0.61 | 1.3e-02 | Click! |
| znf740b | dr10_dc_chr23_-_36319185_36319329 | 0.04 | 8.7e-01 | Click! |
Activity profile of znf740a+znf740b motif
Sorted Z-values of znf740a+znf740b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of znf740a+znf740b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_38834751 | 5.23 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr23_-_511984 | 5.19 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr14_-_9216303 | 4.76 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr2_+_23038657 | 4.74 |
ENSDART00000089012
|
kif1ab
|
kinesin family member 1Ab |
| chr23_+_36002332 | 4.44 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr14_+_21531709 | 4.07 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr11_-_23954234 | 3.93 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr13_-_25067585 | 3.76 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr21_-_26459113 | 3.64 |
ENSDART00000157255
|
cd248b
|
CD248 molecule, endosialin b |
| KN149987v1_-_29629 | 3.59 |
ENSDART00000159555
ENSDART00000168161 |
ENSDARG00000103311
|
ENSDARG00000103311 |
| chr18_+_43041488 | 3.58 |
|
|
|
| chr3_-_60929921 | 3.52 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr2_+_43619752 | 3.41 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| KN150411v1_-_12184 | 3.28 |
|
|
|
| chr22_-_95158 | 3.25 |
|
|
|
| chr25_+_34997593 | 3.18 |
ENSDART00000058443
|
fibina
|
fin bud initiation factor a |
| chr22_+_24131239 | 3.11 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr23_+_32573474 | 3.09 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr3_+_49277125 | 3.08 |
ENSDART00000161724
|
gas7a
|
growth arrest-specific 7a |
| chr23_+_6298911 | 3.07 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr2_-_16896965 | 3.03 |
ENSDART00000022549
|
atp1b3a
|
ATPase, Na+/K+ transporting, beta 3a polypeptide |
| chr11_-_4216204 | 3.02 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr5_-_39910235 | 3.02 |
ENSDART00000146237
ENSDART00000163302 |
fsta
|
follistatin a |
| chr17_+_27383737 | 3.00 |
ENSDART00000156756
|
FP017274.1
|
ENSDARG00000097369 |
| chr14_+_33117964 | 2.99 |
ENSDART00000075278
|
atp1b4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr5_-_70915664 | 2.97 |
|
|
|
| chr7_-_48392300 | 2.94 |
|
|
|
| chr2_-_23348612 | 2.88 |
ENSDART00000110373
|
znf414
|
zinc finger protein 414 |
| chr2_-_39710140 | 2.84 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr1_-_51862897 | 2.83 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr20_+_26639029 | 2.82 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr14_-_46651359 | 2.80 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr18_+_44656323 | 2.76 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr12_+_29994735 | 2.74 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr10_-_43711606 | 2.66 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr10_+_10252074 | 2.61 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr9_-_34146631 | 2.60 |
ENSDART00000177897
|
AL590134.2
|
ENSDARG00000107869 |
| chr21_+_5528438 | 2.56 |
ENSDART00000168158
|
shroom3
|
shroom family member 3 |
| chr9_+_12988528 | 2.52 |
ENSDART00000125961
|
bmpr2b
|
bone morphogenetic protein receptor, type II b (serine/threonine kinase) |
| chr23_+_6043862 | 2.51 |
|
|
|
| chr13_-_508202 | 2.48 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr1_-_37990863 | 2.48 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr17_+_52736192 | 2.42 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr4_-_12863235 | 2.41 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr20_-_35343242 | 2.35 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr16_-_9108885 | 2.35 |
ENSDART00000153785
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr21_+_5483017 | 2.33 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr7_-_69399273 | 2.32 |
ENSDART00000159823
ENSDART00000075178 ENSDART00000168942 ENSDART00000126739 |
tspan5a
|
tetraspanin 5a |
| chr3_-_60930025 | 2.28 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr13_-_31304614 | 2.28 |
|
|
|
| chr17_-_20830287 | 2.28 |
ENSDART00000149630
|
ank3a
|
ankyrin 3a |
| chr20_+_47524781 | 2.27 |
|
|
|
| chr2_-_26872047 | 2.25 |
ENSDART00000138693
|
si:ch211-106k21.5
|
si:ch211-106k21.5 |
| chr2_-_44330405 | 2.22 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr1_-_11286438 | 2.21 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr1_-_50831155 | 2.21 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr13_-_45386047 | 2.20 |
ENSDART00000137566
|
rhd
|
Rh blood group, D antigen |
| chr5_+_42312784 | 2.19 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr8_-_52952226 | 2.19 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr10_-_31619761 | 2.18 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr5_-_13844114 | 2.18 |
|
|
|
| chr2_-_44893769 | 2.15 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr21_-_30144979 | 2.14 |
ENSDART00000065447
|
hnrnph1l
|
heterogeneous nuclear ribonucleoprotein H1, like |
| chr22_+_24131297 | 2.13 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr15_-_5827067 | 2.13 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr11_+_38013238 | 2.10 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr20_+_20772543 | 2.10 |
ENSDART00000152481
|
rtn1b
|
reticulon 1b |
| chr22_+_28496424 | 2.08 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr7_+_71096788 | 2.08 |
ENSDART00000161871
|
sod3a
|
superoxide dismutase 3, extracellular a |
| chr11_-_25803101 | 2.08 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr10_+_22759607 | 2.07 |
|
|
|
| chr10_+_24476213 | 2.07 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_+_32183937 | 2.06 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr9_-_31467299 | 2.06 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr5_-_47519273 | 2.05 |
|
|
|
| chr9_-_32942783 | 2.05 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr5_+_68410884 | 2.04 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr10_+_1812271 | 2.01 |
|
|
|
| KN150342v1_-_59821 | 1.97 |
ENSDART00000163274
|
ENSDARG00000100757
|
ENSDARG00000100757 |
| chr6_-_43679553 | 1.96 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr10_+_9134634 | 1.96 |
ENSDART00000110443
|
fstb
|
follistatin b |
| chr23_+_22763861 | 1.95 |
|
|
|
| chr9_+_7609372 | 1.93 |
|
|
|
| chr7_+_28862183 | 1.92 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr11_+_1996358 | 1.90 |
|
|
|
| chr17_-_20830398 | 1.89 |
ENSDART00000149630
|
ank3a
|
ankyrin 3a |
| chr25_+_36796176 | 1.88 |
ENSDART00000170289
|
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr25_+_36796226 | 1.85 |
ENSDART00000170289
|
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr16_+_22672315 | 1.82 |
ENSDART00000142241
|
she
|
Src homology 2 domain containing E |
| chr6_+_54231519 | 1.80 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr12_-_7820226 | 1.78 |
ENSDART00000149594
ENSDART00000148866 |
ank3b
|
ankyrin 3b |
| chr12_+_13206107 | 1.78 |
ENSDART00000105896
|
atp2a1l
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 like |
| chr13_+_1484997 | 1.76 |
|
|
|
| chr16_+_25845432 | 1.75 |
|
|
|
| chr10_-_43466392 | 1.75 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr7_+_16583234 | 1.75 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr17_-_52735615 | 1.74 |
|
|
|
| chr18_+_35994461 | 1.73 |
|
|
|
| chr9_+_56182756 | 1.73 |
ENSDART00000144757
|
mxra5b
|
matrix-remodelling associated 5b |
| chr23_-_41756325 | 1.68 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr24_-_16994956 | 1.67 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr2_+_24135188 | 1.67 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr14_+_23986703 | 1.66 |
ENSDART00000079164
|
klhl3
|
kelch-like family member 3 |
| chr13_-_37339615 | 1.66 |
ENSDART00000100324
|
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr20_-_53271601 | 1.65 |
|
|
|
| chr4_+_11312432 | 1.65 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr10_-_43466478 | 1.62 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr17_-_37040941 | 1.61 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr13_-_14978365 | 1.61 |
|
|
|
| chr16_-_31476944 | 1.61 |
ENSDART00000167350
|
zgc:194210
|
zgc:194210 |
| chr9_+_5884066 | 1.60 |
ENSDART00000129117
|
pdzk1
|
PDZ domain containing 1 |
| chr5_-_13826859 | 1.58 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr24_-_16995194 | 1.56 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr10_-_43711513 | 1.56 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr20_-_35343057 | 1.56 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr11_+_40945681 | 1.55 |
|
|
|
| chr2_-_4277516 | 1.53 |
|
|
|
| chr16_+_10974655 | 1.53 |
|
|
|
| chr11_+_18823629 | 1.52 |
|
|
|
| chr7_+_34394526 | 1.51 |
|
|
|
| chr9_+_7609527 | 1.51 |
|
|
|
| chr11_-_37921642 | 1.48 |
|
|
|
| chr7_+_890752 | 1.48 |
ENSDART00000111531
|
epdl1
|
ependymin-like 1 |
| chr18_+_17564926 | 1.47 |
|
|
|
| chr24_+_30343717 | 1.47 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr3_+_22412059 | 1.46 |
|
|
|
| chr14_+_31226164 | 1.45 |
ENSDART00000002684
|
ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr12_+_5495284 | 1.45 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr20_+_25441689 | 1.44 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr3_+_40667131 | 1.40 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr1_-_45941709 | 1.39 |
ENSDART00000053232
|
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr3_+_23573114 | 1.38 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr22_-_15182840 | 1.35 |
|
|
|
| chr21_+_18295124 | 1.34 |
ENSDART00000167511
|
ENSDARG00000104101
|
ENSDARG00000104101 |
| chr25_+_33364224 | 1.33 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr3_+_14238188 | 1.33 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr3_+_52901602 | 1.32 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr25_-_7346766 | 1.32 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr18_-_26876598 | 1.32 |
|
|
|
| chr11_-_18542803 | 1.31 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr18_+_21133694 | 1.31 |
ENSDART00000060015
|
chka
|
choline kinase alpha |
| chr22_-_15930756 | 1.29 |
ENSDART00000080047
|
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr5_-_63031745 | 1.29 |
|
|
|
| chr23_-_17543731 | 1.28 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr1_-_57980632 | 1.28 |
ENSDART00000100152
|
sid4
|
secreted immunoglobulin domain 4 |
| chr21_-_14950265 | 1.27 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr4_+_25231425 | 1.27 |
ENSDART00000066934
|
itih5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr21_+_13236509 | 1.27 |
|
|
|
| chr23_+_35984675 | 1.26 |
ENSDART00000053295
|
hoxc10a
|
homeobox C10a |
| chr15_+_3296905 | 1.26 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr19_+_43414160 | 1.26 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr20_-_28739099 | 1.25 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr5_+_36014029 | 1.25 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr12_+_35912088 | 1.23 |
ENSDART00000167873
|
baiap2b
|
BAI1-associated protein 2b |
| chr6_-_43679476 | 1.22 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr25_+_30535271 | 1.20 |
|
|
|
| chr7_-_68417190 | 1.20 |
|
|
|
| chr11_+_38013564 | 1.17 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr1_-_57980586 | 1.16 |
ENSDART00000100152
|
sid4
|
secreted immunoglobulin domain 4 |
| chr18_-_48996702 | 1.16 |
ENSDART00000125432
|
esrrd
|
estrogen-related receptor delta |
| chr16_-_8367090 | 1.15 |
ENSDART00000129068
|
entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr3_+_21059221 | 1.13 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr2_-_54965656 | 1.10 |
|
|
|
| chr5_+_29327479 | 1.10 |
|
|
|
| chr23_+_36002475 | 1.10 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr2_-_6139895 | 1.09 |
ENSDART00000147509
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr2_+_47885551 | 1.09 |
|
|
|
| chr24_+_9911748 | 1.08 |
|
|
|
| chr8_+_8252892 | 1.08 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr17_+_8874210 | 1.08 |
|
|
|
| chr5_+_30099073 | 1.08 |
ENSDART00000012888
|
atp5l
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g |
| chr13_-_508050 | 1.07 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr7_-_28339836 | 1.04 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr1_-_37990935 | 1.01 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr3_-_25239180 | 1.01 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr15_-_10532992 | 1.01 |
ENSDART00000175825
|
tenm4
|
teneurin transmembrane protein 4 |
| chr24_+_30343687 | 0.99 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| KN150342v1_-_60050 | 0.98 |
ENSDART00000163274
|
ENSDARG00000100757
|
ENSDARG00000100757 |
| chr15_+_43882577 | 0.97 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr25_-_10992022 | 0.97 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr22_-_38184317 | 0.97 |
ENSDART00000147694
|
tm4sf4
|
transmembrane 4 L six family member 4 |
| chr16_+_47493635 | 0.97 |
ENSDART00000032309
|
slc25a32b
|
solute carrier family 25 (mitochondrial folate carrier), member 32b |
| chr5_+_29327363 | 0.97 |
|
|
|
| chr5_+_13870125 | 0.96 |
|
|
|
| chr3_-_1421391 | 0.95 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr5_-_18548376 | 0.94 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr23_+_22763789 | 0.94 |
|
|
|
| chr10_+_10252239 | 0.94 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr18_-_34573597 | 0.93 |
ENSDART00000021880
|
ssr3
|
signal sequence receptor, gamma |
| chr5_+_60457842 | 0.92 |
|
|
|
| chr5_-_1498277 | 0.92 |
|
|
|
| chr18_-_35485470 | 0.90 |
ENSDART00000141023
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr22_-_20361311 | 0.90 |
|
|
|
| chr2_+_28906539 | 0.90 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr7_+_20046613 | 0.90 |
ENSDART00000131019
|
acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr21_+_26696853 | 0.89 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr17_-_27247075 | 0.88 |
|
|
|
| chr9_-_53380782 | 0.88 |
ENSDART00000166906
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr8_-_387982 | 0.87 |
|
|
|
| chr21_+_37985816 | 0.86 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 1.2 | 4.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.9 | 3.8 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.8 | 4.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.8 | 2.4 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.7 | 2.2 | GO:0051701 | interaction with host(GO:0051701) |
| 0.7 | 2.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.7 | 5.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.7 | 2.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.6 | 3.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.6 | 2.2 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.5 | 2.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.5 | 3.9 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.4 | 1.3 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.4 | 2.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.4 | 5.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.4 | 3.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.3 | 1.0 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.3 | 1.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.3 | 2.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 2.1 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.3 | 1.7 | GO:0070293 | renal absorption(GO:0070293) |
| 0.3 | 1.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 1.8 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 3.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 0.7 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.2 | 1.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 1.0 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.2 | 4.5 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.2 | 3.0 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 5.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.2 | 2.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 1.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 2.8 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.7 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.0 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 1.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 1.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 2.6 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 4.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 2.4 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 0.3 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 1.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.9 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 3.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 2.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.3 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 1.3 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.1 | 2.2 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 2.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 3.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.9 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 4.7 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 5.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 4.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.7 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 2.4 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.7 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.8 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 1.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.1 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 1.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.5 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 6.9 | GO:0048812 | neuron projection morphogenesis(GO:0048812) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.0 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.6 | 4.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 2.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 4.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 2.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 2.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 2.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 6.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 12.6 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 9.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 4.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 8.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 1.2 | 3.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.5 | 2.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.5 | 2.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.4 | 2.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 1.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 3.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 1.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 3.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.4 | 5.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 4.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 1.0 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.3 | 1.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 1.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 1.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 3.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 3.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.2 | 2.1 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.2 | 3.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 1.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 2.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.8 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 2.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 2.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 5.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 5.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 4.1 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 7.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 4.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 5.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 9.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 6.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 10.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 2.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 5.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 5.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 0.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |