Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
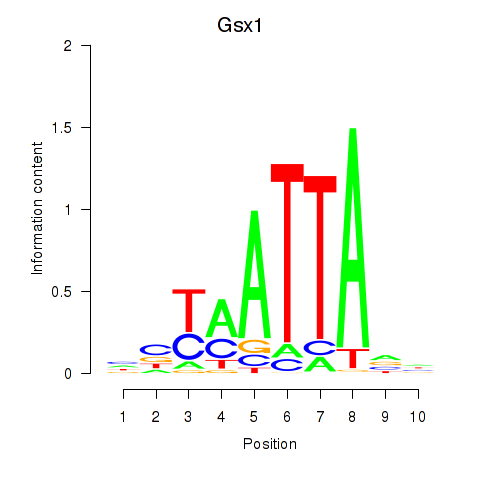
Results for Gsx1_Alx1_Mixl1_Lbx2
Z-value: 0.68
Transcription factors associated with Gsx1_Alx1_Mixl1_Lbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsx1
|
ENSRNOG00000000952 | GS homeobox 1 |
|
Alx1
|
ENSRNOG00000004390 | ALX homeobox 1 |
|
Mixl1
|
ENSRNOG00000003176 | Mix paired-like homeobox 1 |
|
Lbx2
|
ENSRNOG00000052374 | ladybird homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx1 | rn6_v1_chr7_-_44771458_44771458 | -0.49 | 4.1e-01 | Click! |
| Gsx1 | rn6_v1_chr12_-_9607168_9607168 | -0.35 | 5.7e-01 | Click! |
| Mixl1 | rn6_v1_chr13_-_97282299_97282299 | 0.19 | 7.6e-01 | Click! |
Activity profile of Gsx1_Alx1_Mixl1_Lbx2 motif
Sorted Z-values of Gsx1_Alx1_Mixl1_Lbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 2.79 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr10_-_88670430 | 1.07 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr4_-_80395502 | 0.96 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr7_-_28711761 | 0.95 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr17_-_43807540 | 0.79 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr7_+_28654733 | 0.59 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr13_-_32427177 | 0.47 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr5_+_50381244 | 0.46 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr6_+_8886591 | 0.42 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr3_-_66417741 | 0.27 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_-_87232723 | 0.26 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr7_+_136182224 | 0.22 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr15_+_108318664 | 0.21 |
ENSRNOT00000057469
|
Ubac2
|
UBA domain containing 2 |
| chr12_+_47551935 | 0.20 |
ENSRNOT00000056932
|
RGD1560398
|
RGD1560398 |
| chr6_-_125723732 | 0.20 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr8_+_71914867 | 0.19 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr1_-_124803363 | 0.18 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr3_+_5709236 | 0.17 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr9_-_85243001 | 0.17 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr5_-_17061361 | 0.17 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr12_-_17186679 | 0.16 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr2_+_200397967 | 0.16 |
ENSRNOT00000025821
|
Reg4
|
regenerating family member 4 |
| chr15_-_95514259 | 0.16 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr1_+_87224677 | 0.15 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr4_+_6827429 | 0.15 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr4_+_117962319 | 0.15 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr15_+_44441856 | 0.14 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr17_-_61332391 | 0.14 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr19_-_49448072 | 0.14 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr8_+_22559098 | 0.14 |
ENSRNOT00000041091
|
LOC691141
|
hypothetical protein LOC691141 |
| chr2_+_198417619 | 0.14 |
ENSRNOT00000085945
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chrM_+_7758 | 0.13 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr7_-_143967484 | 0.13 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr19_-_37938857 | 0.13 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr10_+_55940533 | 0.13 |
ENSRNOT00000012061
|
RGD1563441
|
similar to RIKEN cDNA A030009H04 |
| chr2_+_158097843 | 0.13 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr4_+_94696965 | 0.13 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr2_-_45518502 | 0.12 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr4_-_157304653 | 0.12 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr6_+_28382962 | 0.12 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr1_-_15374850 | 0.12 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr2_+_51672722 | 0.11 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr4_-_100252755 | 0.11 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr9_-_82146874 | 0.11 |
ENSRNOT00000024190
|
Fev
|
FEV, ETS transcription factor |
| chr11_+_13499164 | 0.11 |
ENSRNOT00000013159
|
LOC680121
|
similar to heat shock protein 8 |
| chr8_+_117117430 | 0.11 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr5_-_17061837 | 0.10 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr2_+_196334626 | 0.10 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr2_+_145174876 | 0.10 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr2_-_138833933 | 0.10 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr7_+_72924799 | 0.10 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr17_-_43640387 | 0.10 |
ENSRNOT00000087731
|
Hist1h1c
|
histone cluster 1 H1 family member c |
| chr3_-_102826379 | 0.10 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr1_-_48825364 | 0.10 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr2_-_185852759 | 0.10 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr8_+_106449321 | 0.10 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr6_-_106971250 | 0.09 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr14_-_44375804 | 0.09 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr5_-_38923095 | 0.09 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr4_+_169147243 | 0.09 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr11_+_46737892 | 0.09 |
ENSRNOT00000052182
|
AABR07033979.1
|
|
| chr20_+_44680449 | 0.09 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr17_-_14627937 | 0.08 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr7_+_44009069 | 0.08 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr17_-_9234766 | 0.08 |
ENSRNOT00000015893
|
AABR07026997.1
|
|
| chrX_-_118998300 | 0.08 |
ENSRNOT00000025575
|
AABR07041089.1
|
|
| chr2_+_80269661 | 0.08 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chrX_+_118197217 | 0.08 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr5_-_154438361 | 0.08 |
ENSRNOT00000085003
|
AC141344.2
|
|
| chrX_-_104932508 | 0.08 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr9_+_10941613 | 0.07 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr6_+_101532518 | 0.07 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr6_-_104290579 | 0.07 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr9_+_20241062 | 0.07 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr11_-_87403367 | 0.07 |
ENSRNOT00000057751
|
Thap7
|
THAP domain containing 7 |
| chr11_-_62067655 | 0.07 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_-_93675991 | 0.07 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr4_+_169161585 | 0.07 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr2_+_225645568 | 0.07 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr20_-_30947484 | 0.06 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr10_+_43601689 | 0.06 |
ENSRNOT00000029238
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr11_+_58624198 | 0.06 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chrX_+_159158194 | 0.06 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr8_-_53816447 | 0.06 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr10_-_87195075 | 0.06 |
ENSRNOT00000014851
|
Krt24
|
keratin 24 |
| chr8_-_78233430 | 0.06 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr1_-_89539210 | 0.06 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr8_+_97291580 | 0.06 |
ENSRNOT00000018794
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr9_-_53315915 | 0.06 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr7_-_143353925 | 0.06 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chr4_+_22898527 | 0.06 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr1_+_85213652 | 0.06 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr5_-_22769907 | 0.06 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr1_-_148443358 | 0.06 |
ENSRNOT00000072973
|
Vbp1
|
VHL binding protein 1 |
| chr1_+_88113445 | 0.06 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr6_+_72124417 | 0.06 |
ENSRNOT00000040548
|
Scfd1
|
sec1 family domain containing 1 |
| chr2_-_256154584 | 0.06 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr2_-_158156444 | 0.06 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr8_-_33121002 | 0.05 |
ENSRNOT00000047211
|
LOC100362981
|
LRRGT00010-like |
| chr8_+_106816152 | 0.05 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chrX_-_14972675 | 0.05 |
ENSRNOT00000079664
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr20_-_45260119 | 0.05 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr9_+_25304876 | 0.05 |
ENSRNOT00000065053
|
Tfap2d
|
transcription factor AP-2 delta |
| chr5_-_115387377 | 0.05 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr3_-_111080705 | 0.05 |
ENSRNOT00000079339
|
Rhov
|
ras homolog family member V |
| chr15_-_37831031 | 0.05 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr9_+_95398237 | 0.05 |
ENSRNOT00000025879
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr15_-_61873406 | 0.05 |
ENSRNOT00000045115
|
LOC100360244
|
LRRGT00053-like |
| chr4_+_155321553 | 0.05 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr6_-_114476723 | 0.05 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr13_-_77959089 | 0.05 |
ENSRNOT00000003586
|
Cacybp
|
calcyclin binding protein |
| chr16_+_50049828 | 0.05 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr9_+_98073038 | 0.05 |
ENSRNOT00000026795
|
Mlph
|
melanophilin |
| chr2_+_54466280 | 0.05 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr2_-_170301348 | 0.05 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr14_+_76732650 | 0.05 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr10_-_87261717 | 0.05 |
ENSRNOT00000015740
|
Krt27
|
keratin 27 |
| chr6_-_7058314 | 0.05 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr10_+_53740841 | 0.05 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr8_+_104106740 | 0.05 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr2_-_46544457 | 0.05 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr2_+_210880777 | 0.05 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr18_+_32336102 | 0.05 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr10_-_29026002 | 0.05 |
ENSRNOT00000005070
|
Pttg1
|
pituitary tumor-transforming 1 |
| chr14_-_16903242 | 0.05 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr1_+_126586543 | 0.04 |
ENSRNOT00000015287
|
Tm2d3
|
TM2 domain containing 3 |
| chr17_+_11683862 | 0.04 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr16_-_73152921 | 0.04 |
ENSRNOT00000048602
|
Zmat4
|
zinc finger, matrin type 4 |
| chrX_-_123980357 | 0.04 |
ENSRNOT00000049435
|
Rhox8
|
reproductive homeobox 8 |
| chr2_+_185846232 | 0.04 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr1_+_22332090 | 0.04 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr20_-_3728844 | 0.04 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr8_+_107229832 | 0.04 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr10_-_40953467 | 0.04 |
ENSRNOT00000092189
|
Glra1
|
glycine receptor, alpha 1 |
| chr18_+_81694808 | 0.04 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chrX_-_77559348 | 0.04 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr7_+_3173287 | 0.04 |
ENSRNOT00000092037
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr3_+_128828331 | 0.04 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chr8_+_47674321 | 0.04 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr3_-_57607683 | 0.04 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr18_-_16542165 | 0.04 |
ENSRNOT00000079381
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr3_+_110855000 | 0.04 |
ENSRNOT00000081613
|
Knl1
|
kinetochore scaffold 1 |
| chr7_-_134560713 | 0.04 |
ENSRNOT00000006621
|
Yaf2
|
YY1 associated factor 2 |
| chr3_-_4341771 | 0.04 |
ENSRNOT00000034694
|
LOC684988
|
similar to ribosomal protein S13 |
| chr5_-_145423172 | 0.04 |
ENSRNOT00000081919
|
Gjb5
|
gap junction protein, beta 5 |
| chr2_-_198016898 | 0.04 |
ENSRNOT00000025523
|
Car14
|
carbonic anhydrase 14 |
| chr6_+_49825469 | 0.04 |
ENSRNOT00000006921
|
Fam150b
|
family with sequence similarity 150, member B |
| chr17_+_44794130 | 0.04 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr18_-_53181503 | 0.04 |
ENSRNOT00000066548
|
Fbn2
|
fibrillin 2 |
| chr13_-_111581018 | 0.04 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr6_+_29977797 | 0.03 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chrX_-_29556610 | 0.03 |
ENSRNOT00000068113
|
Trappc2
|
trafficking protein particle complex 2 |
| chr1_-_102699442 | 0.03 |
ENSRNOT00000056109
|
Tph1
|
tryptophan hydroxylase 1 |
| chr3_-_141411170 | 0.03 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr4_-_138830117 | 0.03 |
ENSRNOT00000008403
|
Il5ra
|
interleukin 5 receptor subunit alpha |
| chr18_-_52047816 | 0.03 |
ENSRNOT00000018368
ENSRNOT00000077491 |
LOC100174910
|
glutaredoxin-like protein |
| chr20_+_3230052 | 0.03 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr10_-_90127600 | 0.03 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr13_-_102857551 | 0.03 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chrX_+_156463953 | 0.03 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr4_+_31704747 | 0.03 |
ENSRNOT00000047850
|
AABR07059675.1
|
|
| chr12_+_4310334 | 0.03 |
ENSRNOT00000041244
|
Vom2r60
|
vomeronasal 2 receptor, 60 |
| chr1_+_87066289 | 0.03 |
ENSRNOT00000027645
|
Capn12
|
calpain 12 |
| chr3_+_173799833 | 0.03 |
ENSRNOT00000081235
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr10_+_54352270 | 0.03 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr2_+_239415046 | 0.03 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr18_-_67224566 | 0.03 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr8_-_83280888 | 0.03 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
| chr2_+_226563050 | 0.03 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr9_-_71852113 | 0.03 |
ENSRNOT00000083263
ENSRNOT00000072983 |
AABR07067896.1
|
|
| chr10_+_5199226 | 0.03 |
ENSRNOT00000003544
|
Dexi
|
dexamethasone-induced transcript |
| chr10_+_91217079 | 0.03 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr4_+_180291389 | 0.03 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr17_-_89163113 | 0.03 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr20_+_19325121 | 0.03 |
ENSRNOT00000058151
|
Phyhipl
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr4_+_158088505 | 0.03 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr12_-_48627297 | 0.03 |
ENSRNOT00000000890
|
Iscu
|
iron-sulfur cluster assembly enzyme |
| chr2_+_240642847 | 0.03 |
ENSRNOT00000079694
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_227932603 | 0.03 |
ENSRNOT00000033795
|
Ms4a6a
|
membrane spanning 4-domains A6A |
| chr17_-_32158538 | 0.03 |
ENSRNOT00000024141
|
Nqo2
|
NAD(P)H quinone dehydrogenase 2 |
| chr7_-_143497108 | 0.03 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr18_+_31444472 | 0.03 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr18_-_24057917 | 0.03 |
ENSRNOT00000023874
|
Rit2
|
Ras-like without CAAX 2 |
| chr3_+_177310753 | 0.03 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr9_-_79630452 | 0.03 |
ENSRNOT00000078125
ENSRNOT00000086044 ENSRNOT00000089283 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr10_-_87286387 | 0.03 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr6_-_127319362 | 0.03 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr2_+_187951344 | 0.03 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chr2_-_158156150 | 0.03 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chrY_-_1398030 | 0.03 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr11_+_45751812 | 0.03 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr15_+_33755478 | 0.03 |
ENSRNOT00000034073
|
Thtpa
|
thiamine triphosphatase |
| chr1_+_240355149 | 0.03 |
ENSRNOT00000018521
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr15_+_56757315 | 0.03 |
ENSRNOT00000078013
|
Esd
|
esterase D |
| chrX_+_131381134 | 0.03 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr8_+_64364741 | 0.03 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr12_+_1460538 | 0.03 |
ENSRNOT00000001444
|
Rfc3
|
replication factor C subunit 3 |
| chr18_+_51523758 | 0.03 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr7_-_29233392 | 0.03 |
ENSRNOT00000064241
|
Spic
|
Spi-C transcription factor |
| chr7_-_73270308 | 0.03 |
ENSRNOT00000007430
|
Rida
|
reactive intermediate imine deaminase A homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsx1_Alx1_Mixl1_Lbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 1.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.6 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.0 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.3 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.1 | 0.2 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 0.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.1 | GO:1903015 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.1 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0071338 | mammary gland bud morphogenesis(GO:0060648) submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.1 | GO:1902276 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) oxygen metabolic process(GO:0072592) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) gamma-aminobutyric acid receptor clustering(GO:0097112) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 2.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |