Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
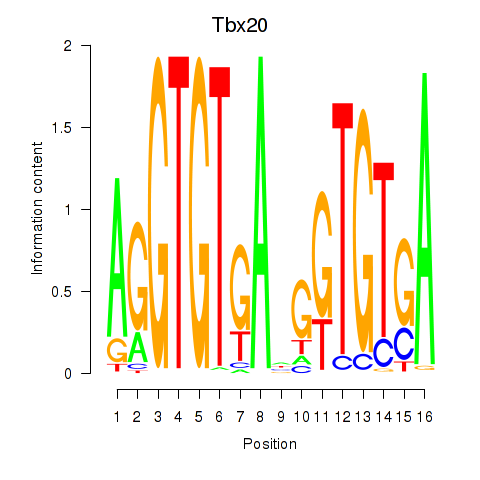
Results for Tbx20
Z-value: 0.39
Transcription factors associated with Tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx20
|
ENSRNOG00000016181 | T-box 20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx20 | rn6_v1_chr8_-_25904564_25904570 | -0.23 | 3.5e-05 | Click! |
Activity profile of Tbx20 motif
Sorted Z-values of Tbx20 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_86579694 | 10.38 |
ENSRNOT00000081209
|
AC119462.1
|
|
| chr10_-_47997796 | 8.46 |
ENSRNOT00000078422
|
Slc5a10
|
solute carrier family 5 member 10 |
| chr19_-_37907714 | 7.96 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr5_+_148661070 | 7.41 |
ENSRNOT00000056229
|
Nkain1
|
Sodium/potassium transporting ATPase interacting 1 |
| chr15_-_42898150 | 7.00 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr9_-_71852113 | 6.88 |
ENSRNOT00000083263
ENSRNOT00000072983 |
AABR07067896.1
|
|
| chr5_-_161035368 | 5.49 |
ENSRNOT00000091640
|
AABR07050321.2
|
|
| chr2_+_2694480 | 5.07 |
ENSRNOT00000083565
|
AABR07072671.1
|
|
| chr6_+_107245820 | 4.93 |
ENSRNOT00000012757
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr1_+_256745288 | 4.26 |
ENSRNOT00000022133
|
Cep55
|
centrosomal protein 55 |
| chr1_-_245817809 | 3.30 |
ENSRNOT00000080607
|
AABR07006672.1
|
|
| chr20_+_44803666 | 3.28 |
ENSRNOT00000057227
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr20_+_29897594 | 3.18 |
ENSRNOT00000057752
|
Vsir
|
V-set immunoregulatory receptor |
| chr6_+_21708487 | 2.50 |
ENSRNOT00000087358
|
AABR07063197.1
|
|
| chr1_+_252944103 | 2.44 |
ENSRNOT00000025770
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr1_-_102845083 | 2.38 |
ENSRNOT00000082289
|
Saa4
|
serum amyloid A4 |
| chr17_-_89163113 | 2.38 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr17_+_20619324 | 2.17 |
ENSRNOT00000079788
|
AABR07027235.1
|
|
| chr1_+_78025995 | 2.16 |
ENSRNOT00000086396
|
Slc8a2
|
solute carrier family 8 member A2 |
| chr20_+_6211420 | 1.99 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr14_-_25585222 | 1.86 |
ENSRNOT00000042106
|
RGD1565048
|
similar to 60S ribosomal protein L9 |
| chr7_+_61165640 | 1.85 |
ENSRNOT00000087955
|
Mdm1
|
Mdm1 nuclear protein |
| chr17_-_14717420 | 1.78 |
ENSRNOT00000071131
|
LOC100910978
|
extracellular matrix protein 2-like |
| chr15_+_51303909 | 1.51 |
ENSRNOT00000085237
ENSRNOT00000058663 |
Loxl2
|
lysyl oxidase-like 2 |
| chr8_-_90984224 | 1.42 |
ENSRNOT00000044931
|
Lca5
|
LCA5, lebercilin |
| chr1_-_141655894 | 1.27 |
ENSRNOT00000082591
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chrX_-_72370044 | 1.25 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr3_-_146812989 | 1.23 |
ENSRNOT00000011315
|
Nanp
|
N-acetylneuraminic acid phosphatase |
| chr4_-_170104933 | 1.15 |
ENSRNOT00000071597
|
Smco3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr4_-_170195107 | 1.12 |
ENSRNOT00000073808
|
LOC100912450
|
uncharacterized LOC100912450 |
| chr17_-_15555919 | 1.09 |
ENSRNOT00000043852
|
Ecm2
|
extracellular matrix protein 2 |
| chrX_+_68752597 | 0.94 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr4_-_170813012 | 0.85 |
ENSRNOT00000073975
|
LOC108348150
|
single-pass membrane and coiled-coil domain-containing protein 3 |
| chr11_-_80826505 | 0.81 |
ENSRNOT00000032383
|
Rtp1
|
receptor (chemosensory) transporter protein 1 |
| chr16_+_19501193 | 0.78 |
ENSRNOT00000085328
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr1_-_141655417 | 0.77 |
ENSRNOT00000068084
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr10_-_44659707 | 0.48 |
ENSRNOT00000002064
|
RGD1559534
|
similar to Alpha enolase (2-phospho-D-glycerate hydro-lyase) |
| chr8_-_20173107 | 0.31 |
ENSRNOT00000045215
|
Olr1163
|
olfactory receptor 1163 |
| chr19_+_22569999 | 0.30 |
ENSRNOT00000022573
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr4_+_1601652 | 0.23 |
ENSRNOT00000072284
|
LOC100912152
|
olfactory receptor 8B3-like |
| chr4_-_165832323 | 0.07 |
ENSRNOT00000007497
|
Tas2r114
|
taste receptor, type 2, member 114 |
| chr20_+_3176107 | 0.07 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx20
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.7 | 3.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.6 | 3.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.4 | 1.2 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.4 | 1.2 | GO:0046380 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.3 | 1.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.3 | 2.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 4.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 8.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 0.8 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 2.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 2.0 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 | 7.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.5 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 2.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.8 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.1 | GO:0032759 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 4.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 7.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 4.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 6.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 1.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.8 | 8.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.6 | 2.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 2.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 1.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 0.8 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.2 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 3.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 7.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 4.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 8.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.0 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 4.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 2.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 3.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |