Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
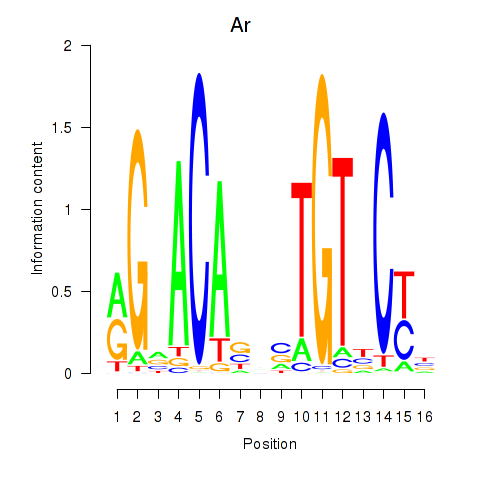
Results for Ar
Z-value: 1.35
Transcription factors associated with Ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ar
|
ENSMUSG00000046532.9 | Ar |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ar | mm39_v1_chrX_+_97192356_97192388 | -0.05 | 7.7e-01 | Click! |
Activity profile of Ar motif
Sorted Z-values of Ar motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ar
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_28453374 | 12.86 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr2_+_58645189 | 4.35 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr4_-_49408040 | 4.18 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr17_-_31363245 | 3.92 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr2_+_58644922 | 3.54 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr12_+_104372962 | 3.17 |
ENSMUST00000021506.6
|
Serpina3n
|
serine (or cysteine) peptidase inhibitor, clade A, member 3N |
| chr14_-_52151537 | 2.87 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr3_-_113371392 | 2.79 |
ENSMUST00000067980.12
|
Amy1
|
amylase 1, salivary |
| chr1_+_88015524 | 2.60 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr3_-_113368407 | 2.59 |
ENSMUST00000106540.8
|
Amy1
|
amylase 1, salivary |
| chr11_+_108286114 | 2.53 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr1_+_88128323 | 2.52 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr3_-_113367891 | 2.28 |
ENSMUST00000142505.9
|
Amy1
|
amylase 1, salivary |
| chr8_-_107792264 | 2.25 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr4_-_137137088 | 2.25 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr3_+_146276147 | 2.06 |
ENSMUST00000199489.5
|
Uox
|
urate oxidase |
| chr7_-_97066937 | 2.05 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr2_+_24970327 | 2.05 |
ENSMUST00000044078.10
ENSMUST00000114380.9 |
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr17_-_12894716 | 2.05 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr7_-_126275529 | 2.04 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr5_+_90608751 | 1.97 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr1_+_172525613 | 1.96 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr6_+_124489364 | 1.91 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr11_+_104467791 | 1.89 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr15_-_82291372 | 1.72 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr17_+_57071765 | 1.67 |
ENSMUST00000007747.10
|
Dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr16_-_18880821 | 1.61 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr1_+_87983099 | 1.57 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_+_104468107 | 1.56 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr2_-_10135449 | 1.55 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr5_-_145656934 | 1.54 |
ENSMUST00000094111.6
|
Cyp3a41a
|
cytochrome P450, family 3, subfamily a, polypeptide 41A |
| chr7_-_99345016 | 1.53 |
ENSMUST00000107086.9
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr5_-_145521533 | 1.50 |
ENSMUST00000075837.8
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr10_+_128104525 | 1.48 |
ENSMUST00000050901.5
|
Apof
|
apolipoprotein F |
| chr7_+_119125546 | 1.46 |
ENSMUST00000207387.2
ENSMUST00000207813.2 |
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chrX_-_154121454 | 1.46 |
ENSMUST00000026328.11
|
Prdx4
|
peroxiredoxin 4 |
| chr9_+_48406706 | 1.46 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr8_+_123912976 | 1.44 |
ENSMUST00000019422.6
|
Dpep1
|
dipeptidase 1 |
| chr15_+_75468473 | 1.42 |
ENSMUST00000189944.7
ENSMUST00000023243.11 |
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr7_+_119125443 | 1.38 |
ENSMUST00000207440.2
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr7_+_119125426 | 1.37 |
ENSMUST00000066465.3
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr15_+_75088445 | 1.37 |
ENSMUST00000055719.8
|
Ly6g2
|
lymphocyte antigen 6 complex, locus G2 |
| chr6_+_138119851 | 1.37 |
ENSMUST00000125810.2
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr1_+_87998487 | 1.35 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr3_+_97536120 | 1.34 |
ENSMUST00000107050.8
ENSMUST00000029729.15 ENSMUST00000107049.2 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr11_-_70537878 | 1.34 |
ENSMUST00000014750.15
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr6_-_72212547 | 1.32 |
ENSMUST00000042646.8
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr10_-_39901249 | 1.31 |
ENSMUST00000163705.3
|
Mfsd4b1
|
major facilitator superfamily domain containing 4B1 |
| chr2_+_118998235 | 1.30 |
ENSMUST00000057454.4
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr15_-_102112159 | 1.27 |
ENSMUST00000229252.2
ENSMUST00000229770.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr8_+_75760301 | 1.19 |
ENSMUST00000165630.3
ENSMUST00000212651.2 ENSMUST00000212388.2 ENSMUST00000212299.2 ENSMUST00000078847.13 ENSMUST00000211869.2 |
Tom1
|
target of myb1 trafficking protein |
| chr17_-_54153367 | 1.18 |
ENSMUST00000023886.7
|
Sult1c2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr11_+_101339233 | 1.18 |
ENSMUST00000010502.13
|
Ifi35
|
interferon-induced protein 35 |
| chr15_+_76579885 | 1.16 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr15_-_79626694 | 1.16 |
ENSMUST00000100439.10
|
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr15_-_79626719 | 1.15 |
ENSMUST00000089311.11
ENSMUST00000046259.14 |
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr17_-_84154173 | 1.12 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr11_+_68989763 | 1.12 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr1_+_87983189 | 1.11 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_-_72892617 | 1.08 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr15_+_75468494 | 1.04 |
ENSMUST00000189874.2
|
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr15_-_102111835 | 1.03 |
ENSMUST00000229043.2
ENSMUST00000230656.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr12_-_113823290 | 1.02 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr15_+_76579960 | 1.00 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr17_-_84154196 | 1.00 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr1_-_180021218 | 1.00 |
ENSMUST00000159914.8
|
Coq8a
|
coenzyme Q8A |
| chr11_+_70538083 | 0.99 |
ENSMUST00000037534.8
|
Rnf167
|
ring finger protein 167 |
| chr7_+_25318829 | 0.99 |
ENSMUST00000077338.12
ENSMUST00000085953.5 |
Dmac2
|
distal membrane arm assembly complex 2 |
| chr1_-_180021039 | 0.98 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr9_-_106562852 | 0.92 |
ENSMUST00000169068.8
|
Tex264
|
testis expressed gene 264 |
| chr4_-_57916283 | 0.92 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr17_+_47022384 | 0.92 |
ENSMUST00000002840.9
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr4_-_44066960 | 0.91 |
ENSMUST00000173234.8
ENSMUST00000173274.2 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr17_+_56563801 | 0.91 |
ENSMUST00000060253.5
|
Fem1a
|
fem 1 homolog a |
| chr15_+_102011352 | 0.90 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr14_+_55829165 | 0.90 |
ENSMUST00000019443.15
|
Rnf31
|
ring finger protein 31 |
| chr15_-_102112657 | 0.88 |
ENSMUST00000231030.2
ENSMUST00000230687.2 ENSMUST00000229514.2 ENSMUST00000229345.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr17_+_34406523 | 0.88 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_+_34406762 | 0.86 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr15_+_102011415 | 0.85 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr12_-_113561594 | 0.84 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr17_-_56381540 | 0.82 |
ENSMUST00000139371.2
|
Ubxn6
|
UBX domain protein 6 |
| chr9_+_121727421 | 0.81 |
ENSMUST00000214340.2
ENSMUST00000050327.5 |
Ackr2
|
atypical chemokine receptor 2 |
| chr2_+_30156523 | 0.79 |
ENSMUST00000091132.13
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr1_+_139429430 | 0.77 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr10_+_87694117 | 0.77 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr3_-_95811993 | 0.76 |
ENSMUST00000147962.3
ENSMUST00000036181.15 |
Car14
|
carbonic anhydrase 14 |
| chr8_+_85809933 | 0.76 |
ENSMUST00000034121.11
|
Man2b1
|
mannosidase 2, alpha B1 |
| chr10_-_78300802 | 0.74 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr15_+_100202061 | 0.72 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr15_-_96917804 | 0.71 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr2_+_25346841 | 0.70 |
ENSMUST00000114265.9
ENSMUST00000102918.3 |
Clic3
|
chloride intracellular channel 3 |
| chr2_-_164621641 | 0.70 |
ENSMUST00000103095.5
|
Tnnc2
|
troponin C2, fast |
| chr11_-_31621863 | 0.68 |
ENSMUST00000058060.14
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr11_-_118021460 | 0.67 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr7_+_101027390 | 0.67 |
ENSMUST00000084895.12
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr2_+_29855572 | 0.67 |
ENSMUST00000113719.9
ENSMUST00000113717.8 ENSMUST00000113741.8 ENSMUST00000100225.9 ENSMUST00000095083.11 ENSMUST00000046257.14 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr16_+_31247562 | 0.67 |
ENSMUST00000115227.10
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr10_-_93375832 | 0.67 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr12_+_84147428 | 0.67 |
ENSMUST00000056822.4
|
Acot6
|
acyl-CoA thioesterase 6 |
| chr7_-_19415301 | 0.66 |
ENSMUST00000150569.9
ENSMUST00000127648.4 ENSMUST00000003071.10 |
Gm44805
Apoc4
|
predicted gene 44805 apolipoprotein C-IV |
| chr6_-_69678271 | 0.66 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr17_-_13159204 | 0.66 |
ENSMUST00000043923.12
|
Acat3
|
acetyl-Coenzyme A acetyltransferase 3 |
| chr19_+_12673147 | 0.65 |
ENSMUST00000025598.10
ENSMUST00000138545.8 ENSMUST00000154822.2 |
Keg1
|
kidney expressed gene 1 |
| chr14_-_33996185 | 0.64 |
ENSMUST00000227006.2
|
Shld2
|
shieldin complex subunit 2 |
| chr11_-_99045894 | 0.64 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr15_+_100202079 | 0.64 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr2_-_62313981 | 0.62 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr6_+_125297596 | 0.61 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr15_+_100202642 | 0.61 |
ENSMUST00000067752.5
ENSMUST00000229588.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr16_-_5040125 | 0.61 |
ENSMUST00000050160.6
|
AU021092
|
expressed sequence AU021092 |
| chr12_-_113700190 | 0.60 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr15_-_74869483 | 0.60 |
ENSMUST00000023248.13
|
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr4_+_116565784 | 0.60 |
ENSMUST00000138305.8
ENSMUST00000125671.8 ENSMUST00000130828.8 |
Ccdc163
|
coiled-coil domain containing 163 |
| chr7_-_46392403 | 0.59 |
ENSMUST00000128088.4
|
Saa1
|
serum amyloid A 1 |
| chr8_+_73072877 | 0.59 |
ENSMUST00000067912.8
|
Klf2
|
Kruppel-like factor 2 (lung) |
| chr4_+_131648509 | 0.58 |
ENSMUST00000238733.2
|
Tmem200b
|
transmembrane protein 200B |
| chr6_+_91133647 | 0.58 |
ENSMUST00000041736.11
|
Hdac11
|
histone deacetylase 11 |
| chr12_-_111874489 | 0.58 |
ENSMUST00000054815.15
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr4_-_44704006 | 0.57 |
ENSMUST00000146335.8
|
Pax5
|
paired box 5 |
| chr15_+_100202021 | 0.57 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr17_-_74257164 | 0.56 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr5_-_24806960 | 0.56 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr10_-_43934774 | 0.56 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr19_+_43770619 | 0.56 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr17_+_34029484 | 0.55 |
ENSMUST00000048560.11
ENSMUST00000172649.8 ENSMUST00000173789.2 |
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr2_-_90900628 | 0.55 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr7_-_99344779 | 0.54 |
ENSMUST00000137914.2
ENSMUST00000207090.2 ENSMUST00000208225.2 |
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr4_+_127019826 | 0.54 |
ENSMUST00000094712.5
|
Tmem35b
|
transmembrane protein 35B |
| chr1_+_172383499 | 0.53 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr4_-_141345549 | 0.52 |
ENSMUST00000053263.9
|
Tmem82
|
transmembrane protein 82 |
| chr17_+_80434874 | 0.52 |
ENSMUST00000039205.11
|
Galm
|
galactose mutarotase |
| chr5_+_25721059 | 0.52 |
ENSMUST00000045016.9
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr1_-_14826208 | 0.52 |
ENSMUST00000027062.7
|
Msc
|
musculin |
| chr9_+_108867633 | 0.51 |
ENSMUST00000112059.10
ENSMUST00000026737.12 |
Shisa5
|
shisa family member 5 |
| chr14_+_70793340 | 0.51 |
ENSMUST00000163060.2
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr17_-_34406193 | 0.51 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr4_-_44703413 | 0.51 |
ENSMUST00000186542.2
|
Pax5
|
paired box 5 |
| chr5_-_108596960 | 0.50 |
ENSMUST00000031455.5
|
Mfsd7a
|
major facilitator superfamily domain containing 7A |
| chr17_+_33743144 | 0.50 |
ENSMUST00000087623.13
ENSMUST00000234715.2 ENSMUST00000234497.2 |
Adamts10
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 |
| chr17_-_23896394 | 0.50 |
ENSMUST00000233428.2
ENSMUST00000233814.2 ENSMUST00000167059.9 ENSMUST00000024698.10 |
Tnfrsf12a
|
tumor necrosis factor receptor superfamily, member 12a |
| chr8_-_65146079 | 0.49 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr11_-_75330415 | 0.49 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr16_-_18884431 | 0.49 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr8_+_84874881 | 0.49 |
ENSMUST00000093375.5
|
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr10_+_21869776 | 0.49 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_-_33500583 | 0.48 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr5_+_8010445 | 0.48 |
ENSMUST00000115421.3
|
Steap4
|
STEAP family member 4 |
| chr11_-_110142565 | 0.48 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr8_+_84728123 | 0.48 |
ENSMUST00000060357.15
ENSMUST00000239176.2 |
1700067K01Rik
|
RIKEN cDNA 1700067K01 gene |
| chr6_+_91134358 | 0.47 |
ENSMUST00000155007.2
|
Hdac11
|
histone deacetylase 11 |
| chr7_-_25089522 | 0.47 |
ENSMUST00000054301.14
|
Lipe
|
lipase, hormone sensitive |
| chr7_-_114162125 | 0.46 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr9_-_57375269 | 0.46 |
ENSMUST00000215059.2
ENSMUST00000046587.8 ENSMUST00000214256.2 |
Scamp5
|
secretory carrier membrane protein 5 |
| chr7_+_6386294 | 0.45 |
ENSMUST00000081022.9
|
Zfp28
|
zinc finger protein 28 |
| chr4_+_116565898 | 0.45 |
ENSMUST00000135499.8
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr4_+_116565706 | 0.45 |
ENSMUST00000030452.13
ENSMUST00000106462.9 |
Ccdc163
|
coiled-coil domain containing 163 |
| chr6_+_55180366 | 0.43 |
ENSMUST00000204842.3
ENSMUST00000053094.8 |
Mindy4
|
MINDY lysine 48 deubiquitinase 4 |
| chr9_+_72892786 | 0.43 |
ENSMUST00000156879.8
|
Ccpg1
|
cell cycle progression 1 |
| chr17_-_24048069 | 0.43 |
ENSMUST00000069579.7
|
Elob
|
elongin B |
| chr8_-_25528972 | 0.43 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr9_+_77848556 | 0.43 |
ENSMUST00000134072.2
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr11_+_59197746 | 0.42 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr19_+_8814782 | 0.42 |
ENSMUST00000171649.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_-_75330302 | 0.42 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr17_+_34879431 | 0.42 |
ENSMUST00000238967.2
|
Tnxb
|
tenascin XB |
| chr13_-_47196633 | 0.42 |
ENSMUST00000021806.11
ENSMUST00000136864.8 |
Tpmt
|
thiopurine methyltransferase |
| chr11_+_94827050 | 0.42 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr5_-_74692327 | 0.41 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr16_-_95993420 | 0.40 |
ENSMUST00000113804.8
ENSMUST00000054855.14 |
Lca5l
|
Leber congenital amaurosis 5-like |
| chr11_-_70242850 | 0.40 |
ENSMUST00000019068.7
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr15_+_89452529 | 0.40 |
ENSMUST00000023295.3
ENSMUST00000230538.2 ENSMUST00000230978.2 |
Acr
|
acrosin prepropeptide |
| chr4_+_128621378 | 0.40 |
ENSMUST00000106079.10
ENSMUST00000133439.8 |
Phc2
|
polyhomeotic 2 |
| chr6_-_68681962 | 0.40 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chrX_-_100838004 | 0.40 |
ENSMUST00000147742.9
|
Gm4779
|
predicted gene 4779 |
| chr12_-_4527138 | 0.39 |
ENSMUST00000085814.5
|
Ncoa1
|
nuclear receptor coactivator 1 |
| chr6_+_91133755 | 0.39 |
ENSMUST00000143621.8
|
Hdac11
|
histone deacetylase 11 |
| chr4_+_149602673 | 0.39 |
ENSMUST00000030839.13
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr10_+_75402090 | 0.38 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr16_-_93400691 | 0.38 |
ENSMUST00000023669.14
ENSMUST00000233931.2 ENSMUST00000154355.3 |
Setd4
|
SET domain containing 4 |
| chr14_+_16728196 | 0.37 |
ENSMUST00000177556.8
|
Gm3373
|
predicted gene 3373 |
| chr3_+_89110223 | 0.37 |
ENSMUST00000077367.11
ENSMUST00000167998.2 |
Gba
|
glucosidase, beta, acid |
| chr13_-_67454476 | 0.36 |
ENSMUST00000049705.8
|
Zfp457
|
zinc finger protein 457 |
| chr12_-_113666198 | 0.36 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr12_-_104120105 | 0.35 |
ENSMUST00000085050.4
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr5_-_31311496 | 0.35 |
ENSMUST00000201353.4
ENSMUST00000200864.4 ENSMUST00000200833.4 ENSMUST00000201491.2 ENSMUST00000200744.4 ENSMUST00000202241.4 ENSMUST00000154241.8 |
Mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr5_+_112436599 | 0.34 |
ENSMUST00000151947.3
|
Tpst2
|
protein-tyrosine sulfotransferase 2 |
| chr9_+_72892693 | 0.34 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr8_+_118428643 | 0.34 |
ENSMUST00000034304.9
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr4_+_116565819 | 0.33 |
ENSMUST00000106463.8
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr6_+_39550791 | 0.33 |
ENSMUST00000140364.8
|
Adck2
|
aarF domain containing kinase 2 |
| chr6_+_39550827 | 0.33 |
ENSMUST00000145788.8
ENSMUST00000051249.13 |
Adck2
|
aarF domain containing kinase 2 |
| chr14_-_70568962 | 0.33 |
ENSMUST00000139284.2
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr3_-_89820451 | 0.32 |
ENSMUST00000029559.7
ENSMUST00000197679.5 |
Il6ra
|
interleukin 6 receptor, alpha |
| chr14_+_16431564 | 0.32 |
ENSMUST00000164139.2
|
Gm8206
|
predicted gene 8206 |
| chr9_-_63056197 | 0.32 |
ENSMUST00000116613.9
|
Skor1
|
SKI family transcriptional corepressor 1 |
| chr7_+_30676465 | 0.32 |
ENSMUST00000058093.6
|
Fam187b
|
family with sequence similarity 187, member B |
| chr15_+_102010632 | 0.32 |
ENSMUST00000229592.2
|
Tns2
|
tensin 2 |
| chr6_+_68657317 | 0.32 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr19_+_8641369 | 0.32 |
ENSMUST00000035444.10
ENSMUST00000163785.2 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 0.8 | 12.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.8 | 9.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.7 | 2.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.6 | 3.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.6 | 1.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.6 | 3.9 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.5 | 2.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.5 | 2.9 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.5 | 2.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.4 | 2.2 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.3 | 1.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.3 | 2.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.3 | 2.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 0.9 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.3 | 2.7 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.3 | 1.5 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.3 | 0.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.2 | 0.7 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.2 | 0.6 | GO:2000412 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 1.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 0.5 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 0.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 2.1 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.2 | 3.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.4 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.1 | 0.5 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.5 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0046340 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 2.0 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.1 | 0.9 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.1 | 0.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.8 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.3 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.1 | 0.3 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.1 | 0.5 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 2.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.5 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.1 | 0.8 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 2.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.6 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.1 | 0.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 2.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 9.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.6 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.4 | GO:1903061 | glucosylceramide catabolic process(GO:0006680) positive regulation of protein lipidation(GO:1903061) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.1 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.1 | 3.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 2.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.1 | 0.4 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) regulation of response to drug(GO:2001023) |
| 0.1 | 0.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.2 | GO:0070948 | neutrophil mediated killing of fungus(GO:0070947) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.1 | GO:0002477 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.4 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.1 | 0.2 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.2 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 1.5 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.1 | 6.2 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.1 | 0.4 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 3.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 7.1 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 1.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.5 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.9 | GO:0006054 | UDP-N-acetylglucosamine metabolic process(GO:0006047) N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.4 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.5 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 6.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability(GO:0035795) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.2 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:1903465 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.0 | 0.1 | GO:1990051 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.5 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 1.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 1.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 2.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) |
| 0.0 | 0.1 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 1.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.6 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.0 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.0 | 0.9 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.3 | 12.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 1.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 7.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 0.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 1.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.4 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 1.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.4 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 11.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 1.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 9.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 4.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.9 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 1.3 | 7.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.1 | 7.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.7 | 2.0 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.6 | 1.7 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.5 | 4.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.5 | 2.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 2.5 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.5 | 1.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 3.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.4 | 1.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.4 | 2.2 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.4 | 2.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 3.0 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.3 | 4.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.7 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.2 | 2.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 9.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 0.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 0.9 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.2 | 4.1 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.2 | 0.7 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 2.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 4.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.4 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 2.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.4 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 1.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.7 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 2.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 3.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.2 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.5 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 6.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 4.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:1990430 | C-X3-C chemokine binding(GO:0019960) extracellular matrix protein binding(GO:1990430) |
| 0.0 | 2.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 2.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 1.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 4.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 5.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 4.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 4.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 3.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 1.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 4.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |