Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Arid5b
Z-value: 0.47
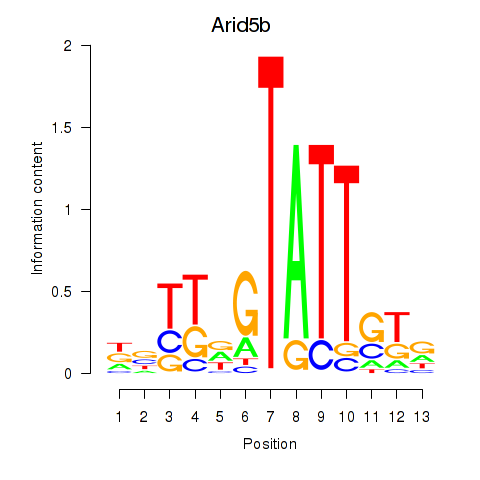
Transcription factors associated with Arid5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5b
|
ENSMUSG00000019947.11 | Arid5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid5b | mm39_v1_chr10_-_68114543_68114570 | -0.19 | 2.7e-01 | Click! |
Activity profile of Arid5b motif
Sorted Z-values of Arid5b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid5b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41435846 | 2.38 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chr6_+_41369290 | 1.77 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chr1_+_174000304 | 1.28 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr18_+_36661198 | 1.02 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr2_-_151586063 | 0.94 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr6_-_125168453 | 0.92 |
ENSMUST00000189959.2
|
Ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr1_-_170755136 | 0.91 |
ENSMUST00000046322.14
ENSMUST00000159171.2 |
Fcrla
|
Fc receptor-like A |
| chr1_+_87603952 | 0.90 |
ENSMUST00000170300.8
ENSMUST00000167032.2 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr17_+_71326510 | 0.84 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr9_-_45896110 | 0.80 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr13_+_28441511 | 0.72 |
ENSMUST00000223428.2
|
Rps18-ps5
|
ribosomal protein S18, pseudogene 5 |
| chr2_-_30720345 | 0.61 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr3_+_103767581 | 0.56 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr17_+_71326542 | 0.56 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr6_+_129374441 | 0.47 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr3_+_95496270 | 0.47 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr3_-_20296337 | 0.47 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr3_+_95496239 | 0.44 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr18_+_42669322 | 0.42 |
ENSMUST00000236418.2
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr9_+_86904067 | 0.42 |
ENSMUST00000168529.9
|
Cyb5r4
|
cytochrome b5 reductase 4 |
| chr7_-_127185512 | 0.41 |
ENSMUST00000205839.2
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr9_-_45895635 | 0.36 |
ENSMUST00000215427.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr6_-_16898440 | 0.35 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chrX_-_156275231 | 0.35 |
ENSMUST00000112529.8
|
Sms
|
spermine synthase |
| chr4_+_132366298 | 0.35 |
ENSMUST00000135299.8
ENSMUST00000020197.14 ENSMUST00000180250.8 ENSMUST00000081726.13 ENSMUST00000079157.11 |
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr16_+_57173456 | 0.34 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr10_+_94412116 | 0.29 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr9_-_42372710 | 0.29 |
ENSMUST00000066179.14
|
Tbcel
|
tubulin folding cofactor E-like |
| chr14_-_86986541 | 0.28 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr7_-_44624165 | 0.27 |
ENSMUST00000212836.2
ENSMUST00000212255.2 ENSMUST00000063761.8 |
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr15_-_36283244 | 0.27 |
ENSMUST00000228358.2
ENSMUST00000022890.10 |
Rnf19a
|
ring finger protein 19A |
| chr10_-_6930376 | 0.27 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_-_19319155 | 0.26 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr6_+_120643323 | 0.25 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chrX_-_100266032 | 0.25 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr13_-_20008397 | 0.24 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr17_+_32671689 | 0.20 |
ENSMUST00000237491.2
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr1_+_180158035 | 0.19 |
ENSMUST00000070181.7
|
Itpkb
|
inositol 1,4,5-trisphosphate 3-kinase B |
| chr12_-_91712783 | 0.19 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2 |
| chr19_-_7688628 | 0.18 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr5_-_31854942 | 0.17 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr10_+_102376109 | 0.17 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr12_-_114321838 | 0.16 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr9_-_45896075 | 0.15 |
ENSMUST00000217636.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr9_-_42368880 | 0.12 |
ENSMUST00000125995.8
|
Tbcel
|
tubulin folding cofactor E-like |
| chr13_+_109822527 | 0.12 |
ENSMUST00000074103.12
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr9_+_104930438 | 0.12 |
ENSMUST00000149243.8
ENSMUST00000035177.15 ENSMUST00000214036.2 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr11_+_11414256 | 0.11 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr11_-_32217547 | 0.11 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr6_+_68414401 | 0.11 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr14_-_26256025 | 0.10 |
ENSMUST00000139075.8
ENSMUST00000102956.8 |
Slmap
|
sarcolemma associated protein |
| chr19_+_13339600 | 0.10 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467 |
| chr3_+_32583681 | 0.10 |
ENSMUST00000147350.8
|
Mfn1
|
mitofusin 1 |
| chr15_-_55411560 | 0.10 |
ENSMUST00000165356.2
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr5_+_136112765 | 0.10 |
ENSMUST00000042135.14
|
Rasa4
|
RAS p21 protein activator 4 |
| chr15_-_44291699 | 0.09 |
ENSMUST00000038719.8
|
Nudcd1
|
NudC domain containing 1 |
| chr15_+_44291470 | 0.09 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr8_-_46542730 | 0.09 |
ENSMUST00000144244.2
|
Snx25
|
sorting nexin 25 |
| chr16_+_58548273 | 0.08 |
ENSMUST00000023426.12
ENSMUST00000162057.8 ENSMUST00000162191.2 |
Cldnd1
|
claudin domain containing 1 |
| chr8_-_83128437 | 0.08 |
ENSMUST00000209573.3
|
Il15
|
interleukin 15 |
| chr17_+_37670473 | 0.08 |
ENSMUST00000178766.3
ENSMUST00000215398.2 |
Olfr104-ps
|
olfactory receptor 104, pseudogene |
| chr7_-_101859033 | 0.07 |
ENSMUST00000211005.2
|
Nup98
|
nucleoporin 98 |
| chr4_-_129556234 | 0.07 |
ENSMUST00000003828.11
|
Kpna6
|
karyopherin (importin) alpha 6 |
| chr7_+_107679062 | 0.06 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr6_-_68857658 | 0.06 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr18_+_77017713 | 0.06 |
ENSMUST00000026487.6
ENSMUST00000136800.2 |
Ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr11_-_17903861 | 0.06 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr6_-_57821483 | 0.05 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr2_+_181023120 | 0.05 |
ENSMUST00000126611.8
|
Lime1
|
Lck interacting transmembrane adaptor 1 |
| chr2_-_34716083 | 0.05 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr16_+_57173632 | 0.05 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr16_-_17540805 | 0.05 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr12_+_103524690 | 0.05 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr8_+_69723732 | 0.04 |
ENSMUST00000212549.2
|
D130040H23Rik
|
RIKEN cDNA D130040H23 gene |
| chr9_-_58065800 | 0.04 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr1_+_31215482 | 0.04 |
ENSMUST00000062560.14
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr2_+_84508734 | 0.04 |
ENSMUST00000102645.4
|
Med19
|
mediator complex subunit 19 |
| chr7_+_33794856 | 0.04 |
ENSMUST00000108083.2
|
Scgb1b30
|
secretoglobin, family 1B, member 30 |
| chr1_+_163607143 | 0.04 |
ENSMUST00000077642.12
ENSMUST00000027877.7 |
Kifap3
|
kinesin-associated protein 3 |
| chr5_+_136112817 | 0.03 |
ENSMUST00000100570.10
|
Rasa4
|
RAS p21 protein activator 4 |
| chr12_-_103956176 | 0.03 |
ENSMUST00000151709.3
ENSMUST00000176246.3 ENSMUST00000074693.13 ENSMUST00000120251.9 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr6_+_97187650 | 0.03 |
ENSMUST00000044681.7
|
Arl6ip5
|
ADP-ribosylation factor-like 6 interacting protein 5 |
| chr8_+_105558204 | 0.03 |
ENSMUST00000059449.7
|
Ces2b
|
carboxyesterase 2B |
| chr9_-_101076198 | 0.03 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr12_-_113617344 | 0.03 |
ENSMUST00000103447.2
|
Ighv2-4
|
immunoglobulin heavy variable V2-4 |
| chr8_-_62576140 | 0.03 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr18_+_37864045 | 0.02 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr10_+_115979787 | 0.02 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr6_+_106095726 | 0.01 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 0.6 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.5 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.4 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.2 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 1.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 4.0 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 1.3 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.0 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.9 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.2 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 1.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.9 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 0.1 | 1.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |