Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Arnt2
Z-value: 0.33
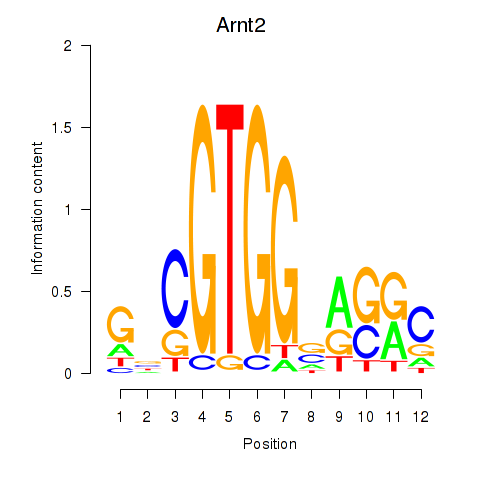
Transcription factors associated with Arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt2
|
ENSMUSG00000015709.10 | Arnt2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arnt2 | mm39_v1_chr7_-_84059321_84059384 | -0.31 | 6.6e-02 | Click! |
Activity profile of Arnt2 motif
Sorted Z-values of Arnt2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Arnt2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_5956251 | 0.35 |
ENSMUST00000060092.13
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr7_-_99276310 | 0.32 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr16_-_4238280 | 0.19 |
ENSMUST00000120080.8
|
Adcy9
|
adenylate cyclase 9 |
| chr1_-_30988772 | 0.19 |
ENSMUST00000238874.2
ENSMUST00000027232.15 ENSMUST00000076587.6 ENSMUST00000233506.2 |
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr15_+_31224616 | 0.17 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr1_-_30988381 | 0.16 |
ENSMUST00000232841.2
|
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr15_+_31224460 | 0.14 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr15_+_31224555 | 0.13 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr7_-_142522454 | 0.12 |
ENSMUST00000121862.3
|
Ascl2
|
achaete-scute family bHLH transcription factor 2 |
| chr10_+_11219117 | 0.11 |
ENSMUST00000069106.5
|
Epm2a
|
epilepsy, progressive myoclonic epilepsy, type 2 gene alpha |
| chr2_+_121859025 | 0.11 |
ENSMUST00000028668.8
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chr5_+_88712840 | 0.10 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr4_+_101353742 | 0.10 |
ENSMUST00000154120.9
ENSMUST00000106930.8 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr6_+_4601124 | 0.08 |
ENSMUST00000141359.2
|
Casd1
|
CAS1 domain containing 1 |
| chrX_-_52358663 | 0.08 |
ENSMUST00000114841.2
ENSMUST00000071023.12 |
Fam122b
|
family with sequence similarity 122, member B |
| chr6_+_4600839 | 0.07 |
ENSMUST00000015333.12
ENSMUST00000181734.8 |
Casd1
|
CAS1 domain containing 1 |
| chr11_+_107438751 | 0.07 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr17_-_88372671 | 0.07 |
ENSMUST00000235112.2
ENSMUST00000005504.15 |
Fbxo11
|
F-box protein 11 |
| chr7_-_45019984 | 0.07 |
ENSMUST00000003971.10
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr11_+_42310557 | 0.07 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr17_-_23964807 | 0.07 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr1_-_135186176 | 0.07 |
ENSMUST00000185752.2
ENSMUST00000003135.14 |
Elf3
|
E74-like factor 3 |
| chr2_-_33261498 | 0.06 |
ENSMUST00000113165.8
|
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr17_-_88372315 | 0.06 |
ENSMUST00000235056.2
ENSMUST00000130379.9 |
Fbxo11
|
F-box protein 11 |
| chr19_+_22425534 | 0.06 |
ENSMUST00000235522.2
ENSMUST00000236372.2 ENSMUST00000238066.2 ENSMUST00000235780.2 ENSMUST00000236804.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr1_+_74627445 | 0.06 |
ENSMUST00000113733.10
ENSMUST00000027358.11 |
Bcs1l
|
BCS1-like (yeast) |
| chr19_+_22425565 | 0.05 |
ENSMUST00000037901.14
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr1_+_59012680 | 0.05 |
ENSMUST00000114296.8
ENSMUST00000027185.11 |
Stradb
|
STE20-related kinase adaptor beta |
| chr5_+_35546363 | 0.05 |
ENSMUST00000172923.2
ENSMUST00000087674.6 |
Hmx1
|
H6 homeobox 1 |
| chr1_-_52856662 | 0.05 |
ENSMUST00000162576.8
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr2_-_33261411 | 0.04 |
ENSMUST00000131298.7
ENSMUST00000091039.5 ENSMUST00000042615.13 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr5_-_135280063 | 0.04 |
ENSMUST00000062572.3
|
Fzd9
|
frizzled class receptor 9 |
| chr4_+_123010035 | 0.03 |
ENSMUST00000102648.6
|
Oxct2b
|
3-oxoacid CoA transferase 2B |
| chr4_-_123217391 | 0.03 |
ENSMUST00000102640.2
|
Oxct2a
|
3-oxoacid CoA transferase 2A |
| chr8_+_84627332 | 0.03 |
ENSMUST00000045393.15
ENSMUST00000132500.8 ENSMUST00000152978.8 |
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr9_+_65278979 | 0.02 |
ENSMUST00000239433.2
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr14_-_63781381 | 0.01 |
ENSMUST00000058679.7
|
Mtmr9
|
myotubularin related protein 9 |
| chr2_-_131404203 | 0.01 |
ENSMUST00000103184.4
|
Adra1d
|
adrenergic receptor, alpha 1d |
| chr9_-_44231526 | 0.01 |
ENSMUST00000214602.2
ENSMUST00000065080.10 |
C2cd2l
|
C2 calcium-dependent domain containing 2-like |
| chr3_+_96604390 | 0.01 |
ENSMUST00000162778.3
ENSMUST00000064900.16 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr2_+_79085844 | 0.01 |
ENSMUST00000099972.5
|
Itga4
|
integrin alpha 4 |
| chr9_-_107971640 | 0.00 |
ENSMUST00000081309.13
ENSMUST00000191985.2 |
Apeh
|
acylpeptide hydrolase |
| chr3_+_96604415 | 0.00 |
ENSMUST00000107077.4
|
Pias3
|
protein inhibitor of activated STAT 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.3 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |