Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
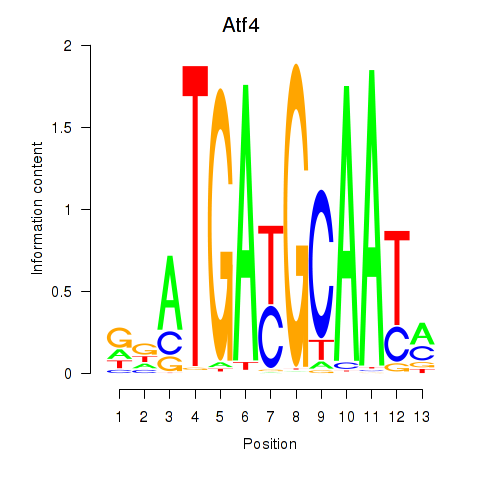
Results for Atf4
Z-value: 2.09
Transcription factors associated with Atf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf4
|
ENSMUSG00000042406.9 | Atf4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf4 | mm39_v1_chr15_+_80139371_80139386 | 0.63 | 3.8e-05 | Click! |
Activity profile of Atf4 motif
Sorted Z-values of Atf4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_36188086 | 13.96 |
ENSMUST00000096089.3
|
Cstdc5
|
cystatin domain containing 5 |
| chr5_-_148336711 | 12.22 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr14_-_56339915 | 11.95 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr16_+_36097505 | 11.18 |
ENSMUST00000042097.11
|
Stfa1
|
stefin A1 |
| chr14_-_69522431 | 9.97 |
ENSMUST00000183882.2
ENSMUST00000037064.5 |
Slc25a37
|
solute carrier family 25, member 37 |
| chr9_-_44255456 | 9.63 |
ENSMUST00000077353.15
|
Hmbs
|
hydroxymethylbilane synthase |
| chr7_+_78563964 | 9.38 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr4_+_43957677 | 9.21 |
ENSMUST00000107855.2
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr5_-_129856237 | 9.20 |
ENSMUST00000118268.9
|
Psph
|
phosphoserine phosphatase |
| chr5_-_129856267 | 9.13 |
ENSMUST00000201394.4
|
Psph
|
phosphoserine phosphatase |
| chr11_-_120534469 | 7.36 |
ENSMUST00000141254.8
ENSMUST00000170556.8 ENSMUST00000151876.8 ENSMUST00000026133.15 ENSMUST00000139706.2 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr14_+_55777807 | 6.94 |
ENSMUST00000226352.2
|
Pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr6_-_83294526 | 6.41 |
ENSMUST00000005810.9
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr7_+_44117444 | 6.13 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr4_+_46450892 | 5.90 |
ENSMUST00000102926.5
|
Anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr4_+_117692583 | 5.74 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_+_44117511 | 5.53 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr19_-_40576897 | 5.46 |
ENSMUST00000025979.13
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr7_+_44117475 | 5.36 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr19_-_40576782 | 5.34 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr7_+_44117404 | 5.25 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr16_-_36154692 | 5.04 |
ENSMUST00000114850.3
|
Cstdc6
|
cystatin domain containing 6 |
| chr4_+_126915104 | 4.88 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr2_+_148640705 | 4.79 |
ENSMUST00000028931.10
ENSMUST00000109947.2 |
Cst8
|
cystatin 8 (cystatin-related epididymal spermatogenic) |
| chr14_-_54655079 | 4.43 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr19_-_40576817 | 4.39 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr12_+_105996961 | 4.08 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr5_-_148336574 | 3.88 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr6_+_55014973 | 3.47 |
ENSMUST00000003572.10
|
Gars
|
glycyl-tRNA synthetase |
| chr7_-_3551003 | 3.45 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr3_+_88523440 | 3.35 |
ENSMUST00000177498.8
ENSMUST00000176500.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr14_-_54655237 | 3.32 |
ENSMUST00000195999.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr19_+_53128901 | 3.29 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr19_-_8690330 | 3.28 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr12_+_73333641 | 3.00 |
ENSMUST00000153941.8
ENSMUST00000122920.8 ENSMUST00000101313.4 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr4_+_129083553 | 2.98 |
ENSMUST00000106054.4
|
Yars
|
tyrosyl-tRNA synthetase |
| chr2_-_3513783 | 2.93 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr6_-_16898440 | 2.82 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr11_-_20282684 | 2.68 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr12_+_85645801 | 2.64 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr9_-_45896663 | 2.58 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr13_+_49835576 | 2.49 |
ENSMUST00000165316.8
ENSMUST00000047363.14 |
Iars
|
isoleucine-tRNA synthetase |
| chr12_+_85646162 | 2.37 |
ENSMUST00000050687.14
|
Jdp2
|
Jun dimerization protein 2 |
| chr19_+_53128861 | 2.35 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr18_-_42395207 | 2.34 |
ENSMUST00000097590.5
|
Lars
|
leucyl-tRNA synthetase |
| chr4_+_124635635 | 2.32 |
ENSMUST00000094782.10
ENSMUST00000153837.8 ENSMUST00000154229.2 |
Inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr2_+_172235702 | 2.24 |
ENSMUST00000099061.9
ENSMUST00000103073.9 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr3_+_88523730 | 2.24 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr11_+_100211363 | 2.20 |
ENSMUST00000152521.2
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr15_-_76594723 | 2.14 |
ENSMUST00000036852.9
|
Recql4
|
RecQ protein-like 4 |
| chr10_+_127126643 | 2.11 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr2_+_172235820 | 2.10 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr18_-_64649497 | 2.05 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr7_-_143153785 | 2.03 |
ENSMUST00000105909.4
ENSMUST00000010899.14 |
Cars
|
cysteinyl-tRNA synthetase |
| chr18_-_64649620 | 2.00 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr7_+_125202653 | 1.96 |
ENSMUST00000206103.2
ENSMUST00000033000.8 |
Il21r
|
interleukin 21 receptor |
| chr18_-_42395131 | 1.95 |
ENSMUST00000236102.2
|
Lars
|
leucyl-tRNA synthetase |
| chr3_-_105839980 | 1.86 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr15_-_76594625 | 1.86 |
ENSMUST00000230544.2
|
Recql4
|
RecQ protein-like 4 |
| chr13_+_49835667 | 1.69 |
ENSMUST00000172254.3
|
Iars
|
isoleucine-tRNA synthetase |
| chr6_-_129484070 | 1.56 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr3_-_96170627 | 1.56 |
ENSMUST00000171473.3
|
H4c14
|
H4 clustered histone 14 |
| chr1_-_78465479 | 1.47 |
ENSMUST00000190441.2
ENSMUST00000170217.8 ENSMUST00000188247.7 ENSMUST00000068333.14 |
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr5_-_31337193 | 1.46 |
ENSMUST00000201428.2
ENSMUST00000201468.4 ENSMUST00000101411.6 |
Gtf3c2
Gm29609
|
general transcription factor IIIC, polypeptide 2, beta predicted gene 29609 |
| chr2_+_163837423 | 1.40 |
ENSMUST00000131288.2
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
| chr7_-_126101484 | 1.33 |
ENSMUST00000166682.9
|
Atxn2l
|
ataxin 2-like |
| chr8_-_106427696 | 1.31 |
ENSMUST00000042608.8
|
Acd
|
adrenocortical dysplasia |
| chr2_+_87853118 | 1.22 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr11_-_98291278 | 1.16 |
ENSMUST00000090827.12
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr12_-_108859123 | 1.16 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr7_-_126101555 | 1.16 |
ENSMUST00000167759.8
|
Atxn2l
|
ataxin 2-like |
| chr7_-_126101245 | 1.16 |
ENSMUST00000179818.3
|
Atxn2l
|
ataxin 2-like |
| chr15_+_82031382 | 1.14 |
ENSMUST00000023100.8
ENSMUST00000229336.2 |
Srebf2
|
sterol regulatory element binding factor 2 |
| chr11_+_67061908 | 0.98 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr11_+_67061837 | 0.96 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr14_-_36857083 | 0.94 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr5_-_31337453 | 0.92 |
ENSMUST00000202639.4
ENSMUST00000088010.12 |
Gtf3c2
|
general transcription factor IIIC, polypeptide 2, beta |
| chr14_-_32407203 | 0.91 |
ENSMUST00000096038.4
|
3425401B19Rik
|
RIKEN cDNA 3425401B19 gene |
| chr3_-_59038031 | 0.83 |
ENSMUST00000091112.6
ENSMUST00000065220.13 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr3_-_89905927 | 0.83 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr4_+_42735912 | 0.83 |
ENSMUST00000107984.2
|
4930578G10Rik
|
RIKEN cDNA 4930578G10 gene |
| chr10_+_101994841 | 0.80 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr1_+_156666485 | 0.76 |
ENSMUST00000111720.2
|
Angptl1
|
angiopoietin-like 1 |
| chr7_-_126224848 | 0.70 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr9_-_15023396 | 0.69 |
ENSMUST00000159985.2
|
Hephl1
|
hephaestin-like 1 |
| chr3_-_123029745 | 0.69 |
ENSMUST00000106426.8
|
Synpo2
|
synaptopodin 2 |
| chr3_-_123029782 | 0.68 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr5_+_145077172 | 0.67 |
ENSMUST00000162594.8
ENSMUST00000162308.8 ENSMUST00000159018.8 ENSMUST00000160075.2 |
Bud31
|
BUD31 homolog |
| chr11_-_98291220 | 0.62 |
ENSMUST00000128897.2
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr14_-_50538979 | 0.61 |
ENSMUST00000216195.2
ENSMUST00000214372.2 ENSMUST00000214756.2 |
Olfr733
|
olfactory receptor 733 |
| chr11_-_115167775 | 0.57 |
ENSMUST00000021078.3
|
Fdxr
|
ferredoxin reductase |
| chr13_-_101829132 | 0.56 |
ENSMUST00000035532.13
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr3_-_127202635 | 0.52 |
ENSMUST00000182959.8
|
Ank2
|
ankyrin 2, brain |
| chr17_+_48769383 | 0.50 |
ENSMUST00000162132.8
|
Unc5cl
|
unc-5 family C-terminal like |
| chr6_+_65019558 | 0.48 |
ENSMUST00000204801.3
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr3_-_89905547 | 0.45 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr3_-_59038539 | 0.44 |
ENSMUST00000198838.2
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr3_-_127202693 | 0.42 |
ENSMUST00000182078.9
|
Ank2
|
ankyrin 2, brain |
| chr15_+_99590098 | 0.41 |
ENSMUST00000228185.2
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr14_-_55108384 | 0.39 |
ENSMUST00000146642.2
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr14_-_50521663 | 0.38 |
ENSMUST00000213701.2
|
Olfr732
|
olfactory receptor 732 |
| chr7_-_126165530 | 0.38 |
ENSMUST00000180459.3
ENSMUST00000032992.7 |
Eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr4_+_138606671 | 0.37 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr4_+_42293921 | 0.37 |
ENSMUST00000238203.2
|
Gm21953
|
predicted gene, 21953 |
| chr2_+_118877610 | 0.37 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr4_+_42735545 | 0.36 |
ENSMUST00000068158.10
|
4930578G10Rik
|
RIKEN cDNA 4930578G10 gene |
| chr10_+_101994719 | 0.34 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr4_-_118036827 | 0.34 |
ENSMUST00000097911.9
|
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr3_-_127202663 | 0.33 |
ENSMUST00000182008.8
ENSMUST00000182547.8 |
Ank2
|
ankyrin 2, brain |
| chr3_+_93227047 | 0.32 |
ENSMUST00000090856.10
ENSMUST00000093774.4 |
Hrnr
|
hornerin |
| chr19_-_50667079 | 0.31 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr1_-_190915441 | 0.29 |
ENSMUST00000027941.14
|
Atf3
|
activating transcription factor 3 |
| chr11_-_79418500 | 0.29 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr5_-_145077048 | 0.27 |
ENSMUST00000031627.9
|
Pdap1
|
PDGFA associated protein 1 |
| chr3_-_59038160 | 0.26 |
ENSMUST00000197841.2
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr6_+_65019574 | 0.26 |
ENSMUST00000031984.9
ENSMUST00000205118.3 |
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_175526152 | 0.26 |
ENSMUST00000094288.10
ENSMUST00000171939.8 |
Wdr64
|
WD repeat domain 64 |
| chr13_-_101829070 | 0.23 |
ENSMUST00000187009.7
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chrX_+_102465616 | 0.22 |
ENSMUST00000182089.2
|
Gm26992
|
predicted gene, 26992 |
| chr2_-_65955338 | 0.22 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chrX_+_163221035 | 0.21 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr9_+_32283779 | 0.20 |
ENSMUST00000047334.10
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr8_-_123202774 | 0.17 |
ENSMUST00000014614.4
|
Rnf166
|
ring finger protein 166 |
| chr2_-_89423470 | 0.14 |
ENSMUST00000217254.2
ENSMUST00000217192.2 ENSMUST00000213221.2 |
Olfr1246
|
olfactory receptor 1246 |
| chr10_-_127358300 | 0.13 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr5_+_93241287 | 0.13 |
ENSMUST00000074733.11
ENSMUST00000201700.4 ENSMUST00000202196.4 ENSMUST00000202308.4 |
Septin11
|
septin 11 |
| chr16_-_29363671 | 0.08 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr10_-_127358231 | 0.06 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_-_92412835 | 0.05 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr13_-_118523760 | 0.02 |
ENSMUST00000022245.10
|
Mrps30
|
mitochondrial ribosomal protein S30 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 3.2 | 9.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 2.3 | 18.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.9 | 5.7 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.6 | 9.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.5 | 7.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 1.4 | 4.3 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 1.4 | 4.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 1.2 | 11.9 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 1.2 | 6.9 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.0 | 4.1 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 1.0 | 7.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.9 | 16.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.9 | 2.7 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.8 | 4.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.7 | 5.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.7 | 3.5 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.7 | 3.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.6 | 4.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.5 | 2.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.4 | 1.3 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.4 | 4.9 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.3 | 1.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 1.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.1 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.2 | 5.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 5.7 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.2 | 2.2 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.2 | 3.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.2 | 1.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 5.0 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.2 | 1.4 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 10.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 22.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 9.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 1.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.3 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 2.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 2.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.7 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 2.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.4 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 2.2 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 5.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.6 | 9.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.5 | 1.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.5 | 4.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 2.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 0.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 9.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 14.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.3 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 5.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 15.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 4.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 17.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 4.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 8.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:0019202 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 3.2 | 9.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 3.1 | 18.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 2.3 | 6.9 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.9 | 9.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.8 | 16.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 1.5 | 7.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 1.4 | 4.3 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 1.4 | 4.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 1.4 | 4.1 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 1.1 | 6.4 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.0 | 4.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.8 | 2.3 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.7 | 4.0 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 0.7 | 10.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.6 | 7.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.6 | 5.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 2.7 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.4 | 2.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 1.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 2.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 22.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 3.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 4.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.8 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 3.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.1 | 1.8 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 11.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 4.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 4.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 6.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 4.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 4.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 6.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 7.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 6.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 3.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 23.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.5 | 16.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.5 | 11.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 5.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 29.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 9.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 1.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 6.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 9.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 1.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 4.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |