Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Batf
Z-value: 0.78
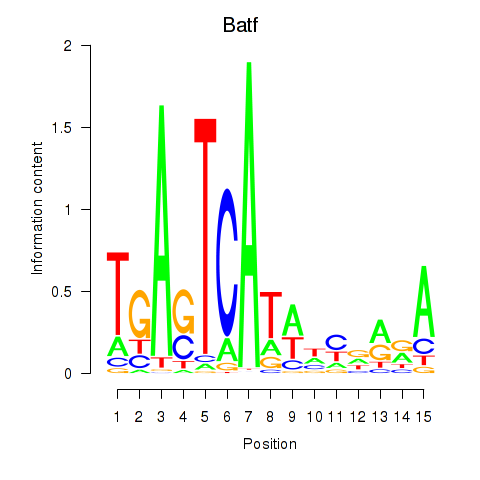
Transcription factors associated with Batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf
|
ENSMUSG00000034266.7 | Batf |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf | mm39_v1_chr12_+_85733415_85733478 | 0.18 | 3.0e-01 | Click! |
Activity profile of Batf motif
Sorted Z-values of Batf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142215027 | 3.59 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr7_-_142215595 | 2.81 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr13_-_21964747 | 1.81 |
ENSMUST00000080511.3
|
H1f5
|
H1.5 linker histone, cluster member |
| chr14_-_32907023 | 0.79 |
ENSMUST00000130509.10
ENSMUST00000061753.15 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr11_-_78950641 | 0.76 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr10_-_62343516 | 0.73 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr3_+_103034843 | 0.72 |
ENSMUST00000172288.3
|
Dennd2c
|
DENN/MADD domain containing 2C |
| chr16_+_35590745 | 0.70 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr1_+_107456731 | 0.69 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr3_-_122828592 | 0.69 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr5_-_44256528 | 0.67 |
ENSMUST00000196178.2
ENSMUST00000197750.5 |
Prom1
|
prominin 1 |
| chr10_+_129219952 | 0.67 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr14_-_32907446 | 0.61 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr2_-_74409225 | 0.57 |
ENSMUST00000134168.2
ENSMUST00000111993.9 ENSMUST00000064503.13 |
Lnpk
|
lunapark, ER junction formation factor |
| chr9_+_108216433 | 0.54 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr7_+_126376353 | 0.51 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr11_+_96941637 | 0.50 |
ENSMUST00000168565.2
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr9_+_108216466 | 0.50 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr11_+_96941420 | 0.48 |
ENSMUST00000090020.13
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr7_-_44861235 | 0.48 |
ENSMUST00000210741.2
ENSMUST00000209466.2 |
Dkkl1
|
dickkopf-like 1 |
| chr19_+_4644425 | 0.47 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr10_+_87926932 | 0.46 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr3_+_14706781 | 0.44 |
ENSMUST00000029071.9
|
Car13
|
carbonic anhydrase 13 |
| chrX_+_149829131 | 0.42 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr13_+_20295672 | 0.41 |
ENSMUST00000180626.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr11_-_78950698 | 0.40 |
ENSMUST00000141409.8
|
Ksr1
|
kinase suppressor of ras 1 |
| chr12_+_83678987 | 0.40 |
ENSMUST00000048155.16
ENSMUST00000182618.8 ENSMUST00000183154.8 ENSMUST00000182036.8 ENSMUST00000182347.8 |
Rbm25
|
RNA binding motif protein 25 |
| chr9_+_113641615 | 0.39 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr12_+_71062733 | 0.38 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr15_+_103362195 | 0.36 |
ENSMUST00000047405.9
|
Nckap1l
|
NCK associated protein 1 like |
| chr10_+_79304565 | 0.35 |
ENSMUST00000233065.2
ENSMUST00000167976.4 |
Vmn2r83
|
vomeronasal 2, receptor 83 |
| chr4_-_117232650 | 0.35 |
ENSMUST00000094853.9
|
Rnf220
|
ring finger protein 220 |
| chr4_-_82777746 | 0.34 |
ENSMUST00000156055.2
ENSMUST00000030110.15 |
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr11_-_97913420 | 0.33 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr18_+_82928959 | 0.31 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr7_-_144305705 | 0.31 |
ENSMUST00000155175.8
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr7_+_101714943 | 0.31 |
ENSMUST00000094130.4
ENSMUST00000084843.10 |
Xndc1
Xntrpc
|
Xrcc1 N-terminal domain containing 1 Xndc1-transient receptor potential cation channel, subfamily C, member 2 readthrough |
| chr3_-_79439181 | 0.30 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr4_+_155953997 | 0.29 |
ENSMUST00000120794.8
ENSMUST00000030901.9 |
Ints11
|
integrator complex subunit 11 |
| chr16_-_19132814 | 0.29 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chr5_+_30218112 | 0.28 |
ENSMUST00000026845.12
ENSMUST00000199183.5 ENSMUST00000195978.5 |
Il6
|
interleukin 6 |
| chr17_-_45903410 | 0.28 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr3_-_63391300 | 0.27 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr3_+_64884839 | 0.27 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr8_-_61407760 | 0.27 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chrX_+_132751729 | 0.26 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr4_+_109092610 | 0.26 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr4_+_109092459 | 0.25 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr7_+_98092628 | 0.25 |
ENSMUST00000098274.5
|
Gucy2d
|
guanylate cyclase 2d |
| chr5_-_6926523 | 0.24 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr5_+_67473069 | 0.24 |
ENSMUST00000202770.2
|
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chrX_-_104919201 | 0.23 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr17_-_56447332 | 0.22 |
ENSMUST00000001256.11
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr16_-_19241884 | 0.22 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr19_-_56810593 | 0.21 |
ENSMUST00000118592.8
|
Ccdc186
|
coiled-coil domain containing 186 |
| chr7_-_85895409 | 0.21 |
ENSMUST00000165771.2
ENSMUST00000233075.2 ENSMUST00000233317.2 ENSMUST00000233312.2 ENSMUST00000233928.2 ENSMUST00000232799.2 ENSMUST00000233744.2 |
Vmn2r76
|
vomeronasal 2, receptor 76 |
| chr11_+_75542902 | 0.21 |
ENSMUST00000102504.10
|
Myo1c
|
myosin IC |
| chr1_+_19279178 | 0.21 |
ENSMUST00000187754.7
|
Tfap2b
|
transcription factor AP-2 beta |
| chr2_+_88470886 | 0.21 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr2_+_86128161 | 0.21 |
ENSMUST00000054746.5
|
Olfr1052
|
olfactory receptor 1052 |
| chr10_+_115979787 | 0.20 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr19_+_29388312 | 0.19 |
ENSMUST00000112576.4
|
Pdcd1lg2
|
programmed cell death 1 ligand 2 |
| chr5_+_31274046 | 0.19 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr17_-_25105277 | 0.19 |
ENSMUST00000234583.2
ENSMUST00000234968.2 ENSMUST00000044252.7 |
Nubp2
|
nucleotide binding protein 2 |
| chrX_-_104918911 | 0.19 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr18_+_37100672 | 0.18 |
ENSMUST00000193777.6
ENSMUST00000193389.2 |
Pcdha6
|
protocadherin alpha 6 |
| chr4_-_59783780 | 0.17 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr18_+_82928782 | 0.17 |
ENSMUST00000235793.2
|
Zfp516
|
zinc finger protein 516 |
| chr18_+_44204457 | 0.17 |
ENSMUST00000068473.4
ENSMUST00000236634.2 |
Spink6
|
serine peptidase inhibitor, Kazal type 6 |
| chr2_-_74409723 | 0.17 |
ENSMUST00000130586.8
|
Lnpk
|
lunapark, ER junction formation factor |
| chr2_-_111843053 | 0.16 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr2_+_88285211 | 0.16 |
ENSMUST00000219871.2
ENSMUST00000213115.3 |
Olfr1183
|
olfactory receptor 1183 |
| chr19_-_4171536 | 0.16 |
ENSMUST00000025767.14
|
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr13_-_32960379 | 0.16 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr7_-_133966588 | 0.16 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr2_+_85648823 | 0.15 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr3_-_37286714 | 0.15 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr8_-_27903965 | 0.15 |
ENSMUST00000033882.10
|
Chrna6
|
cholinergic receptor, nicotinic, alpha polypeptide 6 |
| chr8_-_46577183 | 0.15 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr7_+_115692530 | 0.13 |
ENSMUST00000032899.12
ENSMUST00000106608.8 ENSMUST00000106607.2 |
1110004F10Rik
|
RIKEN cDNA 1110004F10 gene |
| chr18_+_82929037 | 0.13 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr9_+_75682637 | 0.12 |
ENSMUST00000012281.8
|
Bmp5
|
bone morphogenetic protein 5 |
| chr18_+_42408418 | 0.12 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr13_-_65051763 | 0.11 |
ENSMUST00000091554.6
|
Cntnap3
|
contactin associated protein-like 3 |
| chr18_-_43071297 | 0.11 |
ENSMUST00000153737.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr17_-_45903188 | 0.11 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_+_30487322 | 0.10 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr17_+_38082190 | 0.10 |
ENSMUST00000217119.2
|
Olfr122
|
olfactory receptor 122 |
| chr4_-_123466853 | 0.10 |
ENSMUST00000238555.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr1_+_165130192 | 0.10 |
ENSMUST00000111450.3
|
Gpr161
|
G protein-coupled receptor 161 |
| chr9_-_39914761 | 0.10 |
ENSMUST00000215523.2
|
Olfr979
|
olfactory receptor 979 |
| chr13_-_27852017 | 0.09 |
ENSMUST00000006660.7
|
Prl7a2
|
prolactin family 7, subfamily a, member 2 |
| chr1_-_171476559 | 0.09 |
ENSMUST00000111276.9
ENSMUST00000194531.6 |
Slamf7
|
SLAM family member 7 |
| chr3_-_10248979 | 0.09 |
ENSMUST00000029034.9
|
Pmp2
|
peripheral myelin protein 2 |
| chr1_+_177635997 | 0.08 |
ENSMUST00000194528.3
|
Catspere1
|
cation channel sperm associated auxiliary subunit epsilon 1 |
| chr18_+_82929451 | 0.08 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr10_-_117074501 | 0.08 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chr5_+_137759934 | 0.07 |
ENSMUST00000110983.3
ENSMUST00000031738.5 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr18_+_57605525 | 0.07 |
ENSMUST00000209786.2
|
Ctxn3
|
cortexin 3 |
| chrX_+_51880056 | 0.07 |
ENSMUST00000101588.2
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr5_-_86666408 | 0.07 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr18_-_38131766 | 0.07 |
ENSMUST00000236588.2
ENSMUST00000237272.2 ENSMUST00000236134.2 |
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr4_+_58285957 | 0.06 |
ENSMUST00000081919.12
ENSMUST00000177951.8 ENSMUST00000098059.10 ENSMUST00000179951.2 ENSMUST00000102893.10 ENSMUST00000084578.12 ENSMUST00000098057.10 |
Musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr6_-_144993451 | 0.06 |
ENSMUST00000123930.8
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr1_+_134313676 | 0.06 |
ENSMUST00000162187.2
|
Mgat4f
|
MGAT4 family, member F |
| chr10_-_83369994 | 0.06 |
ENSMUST00000020497.14
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr9_+_24009292 | 0.05 |
ENSMUST00000133787.8
ENSMUST00000059650.11 |
Npsr1
|
neuropeptide S receptor 1 |
| chr9_+_37995368 | 0.05 |
ENSMUST00000212502.4
|
Olfr887
|
olfactory receptor 887 |
| chr7_+_118232952 | 0.05 |
ENSMUST00000057320.8
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr2_-_157179344 | 0.05 |
ENSMUST00000109536.8
|
Ghrh
|
growth hormone releasing hormone |
| chr9_+_78522783 | 0.05 |
ENSMUST00000093812.5
|
Cd109
|
CD109 antigen |
| chr7_+_102834996 | 0.05 |
ENSMUST00000218618.2
|
Olfr592
|
olfactory receptor 592 |
| chr11_-_3864664 | 0.04 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr14_+_55459462 | 0.04 |
ENSMUST00000022820.12
|
Dhrs2
|
dehydrogenase/reductase member 2 |
| chr14_+_53048391 | 0.04 |
ENSMUST00000103646.5
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr9_+_106247943 | 0.04 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chrX_-_20955370 | 0.03 |
ENSMUST00000040667.13
|
Zfp300
|
zinc finger protein 300 |
| chr9_+_38026494 | 0.03 |
ENSMUST00000217286.2
|
Olfr889
|
olfactory receptor 889 |
| chr17_+_25105617 | 0.03 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr19_-_46338686 | 0.02 |
ENSMUST00000189008.3
|
2310034G01Rik
|
RIKEN cDNA 2310034G01 gene |
| chr11_-_29775649 | 0.02 |
ENSMUST00000104962.2
|
Fem1al
|
fem-1 homolog A like |
| chr2_+_88505972 | 0.02 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr6_-_122259768 | 0.02 |
ENSMUST00000032207.9
|
Klrg1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr6_-_144993362 | 0.02 |
ENSMUST00000149769.6
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr5_+_8848138 | 0.01 |
ENSMUST00000009058.10
|
Abcb1b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1B |
| chr14_+_54423447 | 0.01 |
ENSMUST00000103709.2
|
Traj32
|
T cell receptor alpha joining 32 |
| chr7_+_126376319 | 0.01 |
ENSMUST00000132643.2
|
Ypel3
|
yippee like 3 |
| chr13_+_27443419 | 0.01 |
ENSMUST00000049463.4
|
Prl3a1
|
prolactin family 3, subfamily a, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 | 1.0 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.2 | 0.7 | GO:0033374 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.2 | 0.5 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.4 | GO:0070358 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.4 | GO:1901581 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 | 0.2 | GO:0097273 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.1 | 1.0 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 0.3 | GO:2000660 | negative regulation of chemokine biosynthetic process(GO:0045079) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.2 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.7 | GO:0009409 | response to cold(GO:0009409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.4 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.4 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.1 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |