Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Batf3
Z-value: 0.75
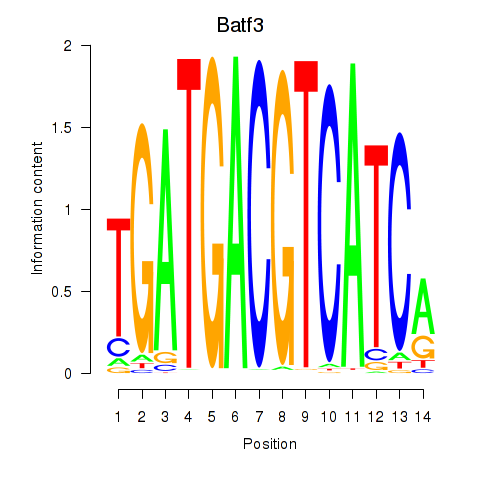
Transcription factors associated with Batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf3
|
ENSMUSG00000026630.10 | Batf3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf3 | mm39_v1_chr1_+_190830522_190830633 | 0.46 | 4.7e-03 | Click! |
Activity profile of Batf3 motif
Sorted Z-values of Batf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_67682312 | 4.20 |
ENSMUST00000224799.2
|
Spire1
|
spire type actin nucleation factor 1 |
| chr6_-_86646118 | 2.60 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr3_+_134918298 | 2.15 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr14_-_87378641 | 1.72 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chr12_+_73333641 | 1.72 |
ENSMUST00000153941.8
ENSMUST00000122920.8 ENSMUST00000101313.4 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr4_+_130253925 | 1.70 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr15_+_79231720 | 1.65 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr15_+_79232137 | 1.61 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr7_+_3706992 | 1.59 |
ENSMUST00000006496.15
ENSMUST00000108623.8 ENSMUST00000139818.2 ENSMUST00000108625.8 |
Rps9
|
ribosomal protein S9 |
| chr6_-_83030759 | 1.49 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr14_+_51045298 | 1.47 |
ENSMUST00000036126.7
|
Parp2
|
poly (ADP-ribose) polymerase family, member 2 |
| chr12_-_69205882 | 1.43 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr13_-_22227114 | 1.37 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr17_-_71833752 | 1.33 |
ENSMUST00000232863.2
ENSMUST00000024851.10 |
Ndc80
|
NDC80 kinetochore complex component |
| chr14_-_51045182 | 1.24 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr2_-_124965537 | 1.16 |
ENSMUST00000142718.8
ENSMUST00000152367.8 ENSMUST00000067780.10 ENSMUST00000147105.8 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr6_-_83031358 | 1.12 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr7_+_97437709 | 1.12 |
ENSMUST00000206984.2
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr13_+_22227359 | 1.08 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr4_-_41275091 | 0.93 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr3_+_40754489 | 0.91 |
ENSMUST00000203295.3
|
Plk4
|
polo like kinase 4 |
| chr2_+_120439858 | 0.86 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr12_+_4132567 | 0.85 |
ENSMUST00000020986.15
ENSMUST00000049584.6 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr5_+_142946098 | 0.83 |
ENSMUST00000031565.15
ENSMUST00000198017.5 |
Fscn1
|
fascin actin-bundling protein 1 |
| chr19_-_61215743 | 0.83 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr6_-_124441731 | 0.83 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr16_+_38383154 | 0.73 |
ENSMUST00000171687.8
ENSMUST00000002924.15 |
Tmem39a
|
transmembrane protein 39a |
| chr4_-_130253703 | 0.58 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr17_-_26727437 | 0.54 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr7_-_79974166 | 0.46 |
ENSMUST00000047362.11
ENSMUST00000121882.8 |
Rccd1
|
RCC1 domain containing 1 |
| chr8_-_61407760 | 0.39 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr5_+_25427860 | 0.39 |
ENSMUST00000045737.14
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr7_-_103778992 | 0.38 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr1_-_126758369 | 0.38 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr5_+_107645626 | 0.36 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr11_+_83553400 | 0.35 |
ENSMUST00000019074.4
|
Ccl4
|
chemokine (C-C motif) ligand 4 |
| chr7_+_90091937 | 0.34 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr9_+_106048116 | 0.33 |
ENSMUST00000020490.13
|
Wdr82
|
WD repeat domain containing 82 |
| chr8_+_106738105 | 0.32 |
ENSMUST00000034375.11
|
Dus2
|
dihydrouridine synthase 2 |
| chr2_-_120439981 | 0.29 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chrX_+_73352694 | 0.27 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr2_-_120439826 | 0.22 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr5_-_130284366 | 0.18 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr17_-_24926589 | 0.17 |
ENSMUST00000126319.8
|
Tbl3
|
transducin (beta)-like 3 |
| chr2_-_120439764 | 0.17 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr4_+_32238712 | 0.16 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr16_+_38383182 | 0.16 |
ENSMUST00000163948.8
|
Tmem39a
|
transmembrane protein 39a |
| chr11_-_69214593 | 0.14 |
ENSMUST00000092973.6
|
Cntrob
|
centrobin, centrosomal BRCA2 interacting protein |
| chr11_+_93935021 | 0.13 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chrX_-_8042129 | 0.08 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr1_+_75145275 | 0.07 |
ENSMUST00000162768.8
ENSMUST00000160439.8 ENSMUST00000027394.12 |
Zfand2b
|
zinc finger, AN1 type domain 2B |
| chr1_-_126758520 | 0.07 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr7_+_16716989 | 0.06 |
ENSMUST00000206129.3
|
Gm42372
|
predicted gene, 42372 |
| chr15_-_76906150 | 0.06 |
ENSMUST00000230031.2
|
Mb
|
myoglobin |
| chr11_+_19874354 | 0.06 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr5_-_124170305 | 0.05 |
ENSMUST00000040967.9
|
Vps37b
|
vacuolar protein sorting 37B |
| chr11_+_93934940 | 0.04 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr10_-_17823736 | 0.04 |
ENSMUST00000037879.8
|
Heca
|
hdc homolog, cell cycle regulator |
| chr14_-_55950939 | 0.03 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr9_-_119151428 | 0.03 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr11_-_70128462 | 0.03 |
ENSMUST00000100950.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr7_+_138448061 | 0.02 |
ENSMUST00000041097.13
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr17_-_36291087 | 0.01 |
ENSMUST00000055454.14
|
Prr3
|
proline-rich polypeptide 3 |
| chr11_+_93935066 | 0.00 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.9 | 2.6 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.5 | 1.5 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.3 | 0.9 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 2.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 1.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 1.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.9 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 1.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.8 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.4 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.8 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 3.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 2.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 0.8 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 2.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 4.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.3 | 2.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 0.8 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 3.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 2.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 4.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.7 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 3.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |