Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
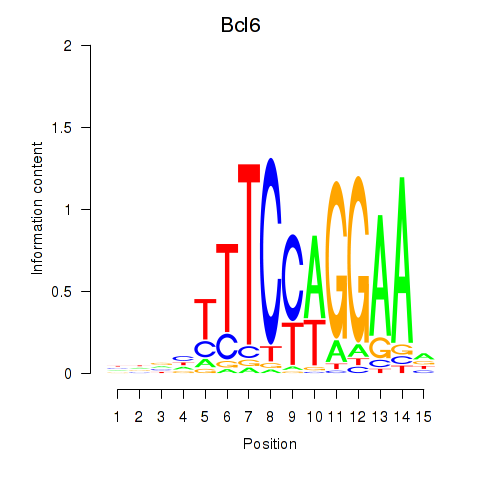
Results for Bcl6
Z-value: 1.32
Transcription factors associated with Bcl6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bcl6
|
ENSMUSG00000022508.6 | Bcl6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl6 | mm39_v1_chr16_-_23807602_23807602 | -0.75 | 1.3e-07 | Click! |
Activity profile of Bcl6 motif
Sorted Z-values of Bcl6 motif
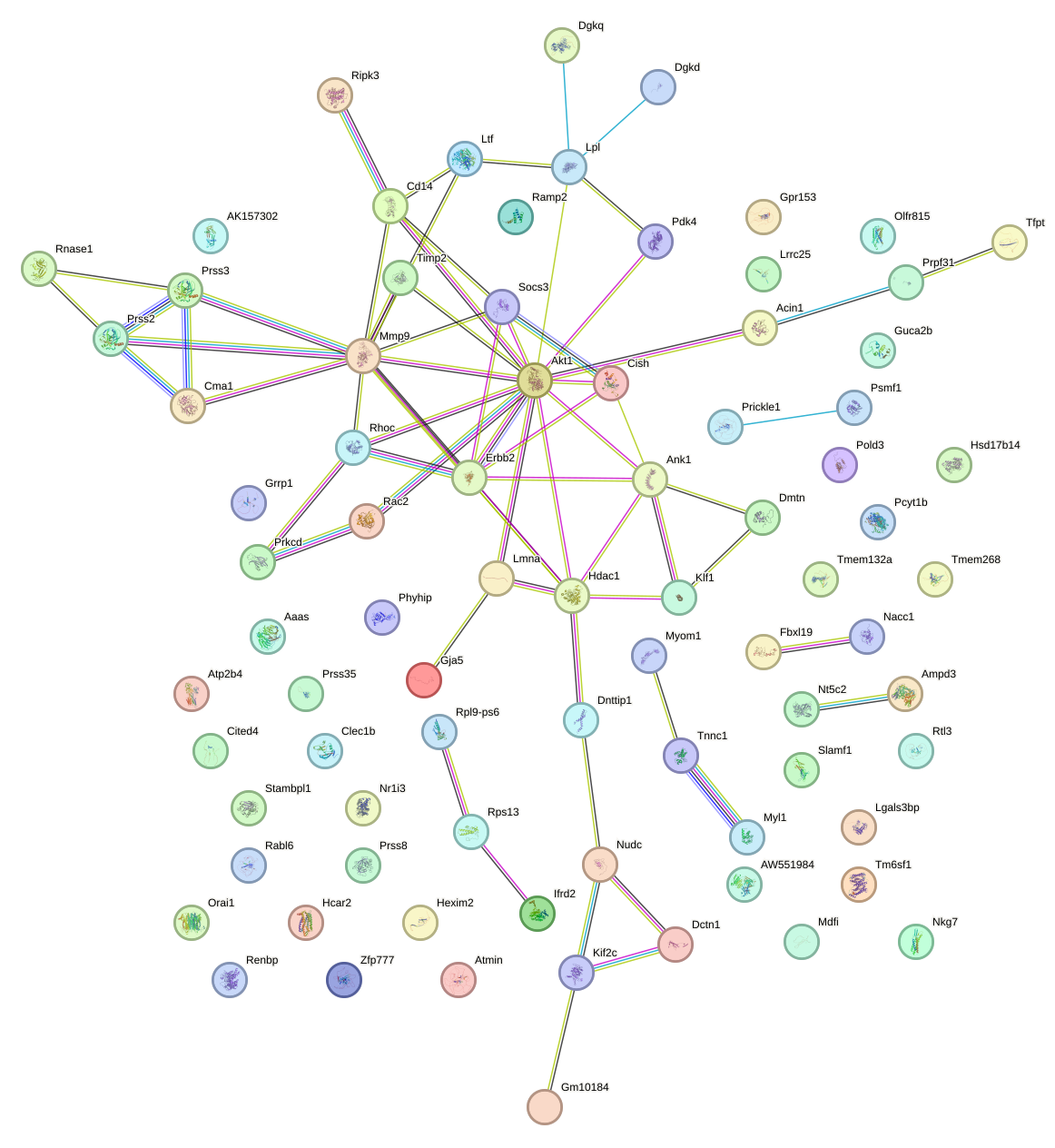
Network of associatons between targets according to the STRING database.
First level regulatory network of Bcl6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110867807 | 10.58 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr8_+_85628557 | 6.72 |
ENSMUST00000067060.10
ENSMUST00000239392.2 |
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chr8_+_23525101 | 6.17 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr4_+_120523758 | 6.04 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr4_+_114916703 | 6.03 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr2_+_164790139 | 5.84 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr7_-_142215027 | 5.28 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr6_+_41498716 | 4.84 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr14_-_70864448 | 4.56 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr14_-_70864666 | 4.27 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr7_+_43086432 | 3.83 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chrX_-_72974357 | 3.29 |
ENSMUST00000155597.2
ENSMUST00000114379.8 |
Renbp
|
renin binding protein |
| chr7_+_43086554 | 3.11 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr6_-_41354538 | 3.07 |
ENSMUST00000096003.7
|
Prss3
|
protease, serine 3 |
| chr1_-_133681419 | 3.03 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chrX_-_72974440 | 2.93 |
ENSMUST00000116578.8
|
Renbp
|
renin binding protein |
| chr3_-_14843512 | 2.85 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr14_+_70694887 | 2.80 |
ENSMUST00000003561.10
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chr4_-_117035922 | 2.75 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr14_-_51384236 | 2.67 |
ENSMUST00000080126.4
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr15_-_78456898 | 2.65 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr3_-_88417251 | 2.34 |
ENSMUST00000149068.2
|
Lmna
|
lamin A |
| chr14_-_56026266 | 2.28 |
ENSMUST00000168716.8
ENSMUST00000178399.3 ENSMUST00000022830.14 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr1_+_135656885 | 2.20 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr19_-_32443978 | 2.19 |
ENSMUST00000078034.5
|
Rpl9-ps6
|
ribosomal protein L9, pseudogene 6 |
| chr13_+_21679387 | 2.19 |
ENSMUST00000104942.2
|
AK157302
|
cDNA sequence AK157302 |
| chr7_+_110372860 | 2.13 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr8_+_71069476 | 2.13 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr11_-_106084334 | 1.97 |
ENSMUST00000007444.14
ENSMUST00000152008.2 ENSMUST00000103072.10 ENSMUST00000106867.2 |
Strada
|
STE20-related kinase adaptor alpha |
| chr4_-_63965161 | 1.95 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr9_+_107173907 | 1.95 |
ENSMUST00000168260.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr7_+_110367375 | 1.89 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr4_+_114945905 | 1.85 |
ENSMUST00000171877.8
ENSMUST00000177647.8 ENSMUST00000106548.9 ENSMUST00000030488.3 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr6_+_129374441 | 1.83 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr14_-_56181993 | 1.75 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr6_+_68247469 | 1.73 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr9_-_39515420 | 1.73 |
ENSMUST00000042485.11
ENSMUST00000141370.8 |
AW551984
|
expressed sequence AW551984 |
| chr6_+_129374260 | 1.71 |
ENSMUST00000032262.14
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr2_-_151586063 | 1.70 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr14_-_32907023 | 1.67 |
ENSMUST00000130509.10
ENSMUST00000061753.15 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr7_+_3632982 | 1.64 |
ENSMUST00000179769.8
ENSMUST00000008517.13 |
Prpf31
|
pre-mRNA processing factor 31 |
| chr4_+_152358648 | 1.63 |
ENSMUST00000105650.8
ENSMUST00000105651.8 |
Gpr153
|
G protein-coupled receptor 153 |
| chr3_-_144412394 | 1.62 |
ENSMUST00000200532.2
|
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr2_-_25498459 | 1.60 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr17_-_90217868 | 1.58 |
ENSMUST00000086423.6
|
Gm10184
|
predicted pseudogene 10184 |
| chr17_+_71326510 | 1.58 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chrX_+_92718695 | 1.57 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr3_+_104696108 | 1.56 |
ENSMUST00000002303.12
|
Rhoc
|
ras homolog family member C |
| chr1_-_66984178 | 1.54 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr7_-_115933367 | 1.51 |
ENSMUST00000205490.2
ENSMUST00000170953.3 |
Rps13
|
ribosomal protein S13 |
| chr19_+_34194990 | 1.49 |
ENSMUST00000119603.2
|
Stambpl1
|
STAM binding protein like 1 |
| chrX_-_105884178 | 1.44 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chr7_+_81508741 | 1.44 |
ENSMUST00000041890.8
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr8_+_3565377 | 1.38 |
ENSMUST00000111070.4
ENSMUST00000004681.14 ENSMUST00000208310.2 |
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr19_-_46950355 | 1.31 |
ENSMUST00000236501.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr4_+_63478478 | 1.25 |
ENSMUST00000080336.4
|
Tmem268
|
transmembrane protein 268 |
| chr9_+_107174081 | 1.24 |
ENSMUST00000167072.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr12_-_112637998 | 1.23 |
ENSMUST00000128300.9
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr4_+_63478454 | 1.22 |
ENSMUST00000124332.8
ENSMUST00000150360.8 |
Tmem268
|
transmembrane protein 268 |
| chr8_-_85414528 | 1.21 |
ENSMUST00000001975.6
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr6_+_83162928 | 1.20 |
ENSMUST00000113907.2
|
Dctn1
|
dynactin 1 |
| chr1_+_87780985 | 1.18 |
ENSMUST00000027517.14
|
Dgkd
|
diacylglycerol kinase, delta |
| chr9_+_86625694 | 1.18 |
ENSMUST00000179574.2
ENSMUST00000036426.13 |
Prss35
|
protease, serine 35 |
| chr10_-_129738595 | 1.18 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr6_-_5496261 | 1.18 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr4_-_133981387 | 1.17 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr1_-_66984521 | 1.15 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr7_-_127529238 | 1.14 |
ENSMUST00000032988.10
ENSMUST00000206124.2 |
Prss8
|
protease, serine 8 (prostasin) |
| chr3_+_96939732 | 1.13 |
ENSMUST00000132256.8
ENSMUST00000072600.7 |
Gja5
|
gap junction protein, alpha 5 |
| chr15_-_102259158 | 1.10 |
ENSMUST00000231061.2
ENSMUST00000041208.9 |
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr9_+_72952115 | 1.09 |
ENSMUST00000184146.8
ENSMUST00000034722.5 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr11_+_103024128 | 1.08 |
ENSMUST00000107037.8
ENSMUST00000124928.2 ENSMUST00000062530.5 |
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr7_-_99770653 | 1.08 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr1_-_167112784 | 1.07 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr8_+_117670126 | 1.06 |
ENSMUST00000109099.4
|
Atmin
|
ATM interactor |
| chr8_+_69333143 | 1.05 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr11_+_101136821 | 1.01 |
ENSMUST00000129680.8
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr6_+_68098030 | 0.99 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr18_-_36859732 | 0.99 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chrX_-_36255377 | 0.98 |
ENSMUST00000152291.3
|
Septin6
|
septin 6 |
| chr19_-_10533088 | 0.97 |
ENSMUST00000059582.9
ENSMUST00000154383.2 |
Tmem216
|
transmembrane protein 216 |
| chr9_+_107468146 | 0.94 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr15_-_93417380 | 0.94 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr1_+_171594690 | 0.93 |
ENSMUST00000015460.5
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr5_-_124003553 | 0.93 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr7_+_127344942 | 0.92 |
ENSMUST00000189562.7
ENSMUST00000186116.7 |
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr11_-_118292678 | 0.91 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr15_+_79025523 | 0.91 |
ENSMUST00000040077.8
|
Polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr17_+_71326542 | 0.90 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr4_-_129436465 | 0.90 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr8_-_85414220 | 0.89 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr8_+_3565401 | 0.88 |
ENSMUST00000207146.2
ENSMUST00000208002.2 |
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr14_-_30329765 | 0.87 |
ENSMUST00000112207.8
ENSMUST00000112206.8 ENSMUST00000112202.8 ENSMUST00000112203.2 |
Prkcd
|
protein kinase C, delta |
| chr7_-_3632826 | 0.86 |
ENSMUST00000205596.2
ENSMUST00000155592.8 ENSMUST00000108641.10 |
Tfpt
|
TCF3 (E2A) fusion partner |
| chr2_-_119617985 | 0.85 |
ENSMUST00000110793.8
ENSMUST00000099529.9 ENSMUST00000048493.12 |
Rpap1
|
RNA polymerase II associated protein 1 |
| chr2_+_164587948 | 0.84 |
ENSMUST00000109327.4
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr6_-_57827328 | 0.83 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr4_-_119515978 | 0.82 |
ENSMUST00000106309.9
ENSMUST00000044426.8 |
Guca2b
|
guanylate cyclase activator 2b (retina) |
| chr11_-_118292758 | 0.82 |
ENSMUST00000043722.10
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr11_+_98303287 | 0.81 |
ENSMUST00000058295.6
|
Erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr11_-_118246566 | 0.81 |
ENSMUST00000155707.3
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr14_-_54891073 | 0.80 |
ENSMUST00000126166.8
ENSMUST00000141453.8 ENSMUST00000150371.8 ENSMUST00000123875.2 ENSMUST00000022794.14 ENSMUST00000148754.10 |
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr17_-_48145466 | 0.79 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr7_-_99770280 | 0.76 |
ENSMUST00000208184.2
|
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr7_+_45204317 | 0.75 |
ENSMUST00000107752.12
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr19_+_12647803 | 0.74 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr4_-_133273243 | 0.73 |
ENSMUST00000030665.7
|
Nudc
|
nudC nuclear distribution protein |
| chr8_-_105350898 | 0.73 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr6_-_48025845 | 0.73 |
ENSMUST00000095944.10
|
Zfp777
|
zinc finger protein 777 |
| chr3_-_126918491 | 0.73 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain |
| chr12_-_114286421 | 0.72 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr19_-_10847121 | 0.72 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr11_-_117859997 | 0.72 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chr5_+_123153072 | 0.71 |
ENSMUST00000051016.5
ENSMUST00000121652.8 |
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chrX_+_162923474 | 0.71 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr1_-_134883645 | 0.70 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_34153328 | 0.69 |
ENSMUST00000056355.9
|
Nat8l
|
N-acetyltransferase 8-like |
| chr9_+_50664288 | 0.69 |
ENSMUST00000214962.2
ENSMUST00000216755.2 |
Cryab
|
crystallin, alpha B |
| chr4_-_130068484 | 0.69 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr15_-_79025387 | 0.68 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr6_+_68026941 | 0.68 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr2_+_150590956 | 0.68 |
ENSMUST00000094467.6
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 |
| chr6_+_28475099 | 0.67 |
ENSMUST00000168362.2
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr5_-_137145030 | 0.67 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr4_-_135000109 | 0.66 |
ENSMUST00000037099.9
|
Clic4
|
chloride intracellular channel 4 (mitochondrial) |
| chr9_+_50664207 | 0.66 |
ENSMUST00000034562.9
|
Cryab
|
crystallin, alpha B |
| chr8_-_11329656 | 0.65 |
ENSMUST00000208095.2
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr11_+_96242422 | 0.65 |
ENSMUST00000100523.7
|
Hoxb2
|
homeobox B2 |
| chr1_+_170846482 | 0.64 |
ENSMUST00000078825.5
|
Fcgr4
|
Fc receptor, IgG, low affinity IV |
| chr8_-_96580129 | 0.64 |
ENSMUST00000212628.2
ENSMUST00000040481.4 ENSMUST00000212270.2 |
Slc38a7
|
solute carrier family 38, member 7 |
| chr7_-_29898236 | 0.64 |
ENSMUST00000001845.13
|
Capns1
|
calpain, small subunit 1 |
| chr3_+_95406567 | 0.63 |
ENSMUST00000015664.5
|
Ctsk
|
cathepsin K |
| chr7_+_24776630 | 0.63 |
ENSMUST00000179556.2
ENSMUST00000053410.11 |
Zfp574
|
zinc finger protein 574 |
| chr4_-_32602760 | 0.63 |
ENSMUST00000219644.2
ENSMUST00000056517.3 |
Gja10
|
gap junction protein, alpha 10 |
| chr14_-_32907446 | 0.62 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr10_-_79940168 | 0.61 |
ENSMUST00000219260.2
|
Sbno2
|
strawberry notch 2 |
| chr19_-_10533562 | 0.60 |
ENSMUST00000025569.9
|
Tmem216
|
transmembrane protein 216 |
| chr17_+_28910302 | 0.60 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr19_-_40260286 | 0.59 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr15_-_80989200 | 0.56 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr19_-_5345423 | 0.56 |
ENSMUST00000235182.2
ENSMUST00000025774.11 |
Sf3b2
|
splicing factor 3b, subunit 2 |
| chr8_+_107237483 | 0.56 |
ENSMUST00000080797.8
|
Cdh3
|
cadherin 3 |
| chr17_+_28910393 | 0.56 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr11_+_68582923 | 0.55 |
ENSMUST00000018887.15
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr18_+_44403169 | 0.55 |
ENSMUST00000042747.4
|
Npy6r
|
neuropeptide Y receptor Y6 |
| chr5_+_86219593 | 0.55 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr4_-_43710231 | 0.54 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr14_-_55950939 | 0.53 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr12_-_114226570 | 0.52 |
ENSMUST00000103479.4
ENSMUST00000195619.2 |
Ighv3-5
|
immunoglobulin heavy variable 3-5 |
| chr6_-_30873669 | 0.51 |
ENSMUST00000048774.13
ENSMUST00000166192.7 |
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr11_-_99121822 | 0.50 |
ENSMUST00000103133.4
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr13_-_63006176 | 0.50 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr8_-_105350881 | 0.49 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr8_+_73197718 | 0.48 |
ENSMUST00000064853.13
ENSMUST00000121902.2 |
1700030K09Rik
|
RIKEN cDNA 1700030K09 gene |
| chr9_-_58066484 | 0.47 |
ENSMUST00000041477.15
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr13_+_23940964 | 0.47 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr11_+_55104609 | 0.47 |
ENSMUST00000108867.2
|
Slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr16_-_4698148 | 0.46 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr18_+_37453427 | 0.46 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr18_+_61178211 | 0.45 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr19_-_40260060 | 0.45 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr16_-_36605286 | 0.45 |
ENSMUST00000168279.2
ENSMUST00000164579.9 ENSMUST00000023616.11 |
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr11_-_55310724 | 0.45 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr17_+_29900144 | 0.43 |
ENSMUST00000127695.2
|
Cmtr1
|
cap methyltransferase 1 |
| chr19_+_46329552 | 0.43 |
ENSMUST00000128041.8
|
Mfsd13a
|
major facilitator superfamily domain containing 13a |
| chr1_+_125604706 | 0.43 |
ENSMUST00000027581.7
|
Gpr39
|
G protein-coupled receptor 39 |
| chr5_-_135601887 | 0.43 |
ENSMUST00000004936.10
ENSMUST00000201401.2 |
Ccl24
|
chemokine (C-C motif) ligand 24 |
| chr10_-_78248771 | 0.42 |
ENSMUST00000062678.11
|
Rrp1
|
ribosomal RNA processing 1 |
| chr11_+_76792977 | 0.42 |
ENSMUST00000102495.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr3_+_138232176 | 0.38 |
ENSMUST00000200020.5
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr19_-_43974990 | 0.37 |
ENSMUST00000026210.5
|
Cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr9_-_35179042 | 0.34 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chr5_+_20433169 | 0.33 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_-_63482668 | 0.32 |
ENSMUST00000118022.8
|
Gata4
|
GATA binding protein 4 |
| chr5_-_6926523 | 0.32 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr9_+_122980006 | 0.31 |
ENSMUST00000026890.6
|
Clec3b
|
C-type lectin domain family 3, member b |
| chr14_-_63482720 | 0.31 |
ENSMUST00000067417.10
|
Gata4
|
GATA binding protein 4 |
| chr11_+_83743746 | 0.31 |
ENSMUST00000108113.3
|
Hnf1b
|
HNF1 homeobox B |
| chr7_+_126575510 | 0.29 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr17_+_21031817 | 0.29 |
ENSMUST00000232810.2
ENSMUST00000233712.2 ENSMUST00000232852.2 |
Vmn1r229
|
vomeronasal 1 receptor 229 |
| chr12_-_115884332 | 0.28 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr12_-_114263874 | 0.27 |
ENSMUST00000103482.2
ENSMUST00000194159.2 |
Ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr10_+_99851679 | 0.27 |
ENSMUST00000130190.8
ENSMUST00000218200.2 ENSMUST00000020129.8 |
Kitl
|
kit ligand |
| chr9_-_100453102 | 0.27 |
ENSMUST00000093792.4
|
Slc35g2
|
solute carrier family 35, member G2 |
| chr2_-_87466089 | 0.26 |
ENSMUST00000090711.3
|
Olfr1132
|
olfactory receptor 1132 |
| chr5_-_103777145 | 0.26 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr4_+_111272156 | 0.26 |
ENSMUST00000030274.7
|
Bend5
|
BEN domain containing 5 |
| chr1_+_180771600 | 0.26 |
ENSMUST00000161523.8
|
Tmem63a
|
transmembrane protein 63a |
| chr3_-_95662134 | 0.25 |
ENSMUST00000198289.5
ENSMUST00000196868.5 ENSMUST00000074339.13 ENSMUST00000163530.8 ENSMUST00000029752.15 ENSMUST00000195929.5 ENSMUST00000199570.2 ENSMUST00000098857.9 |
Tars2
|
threonyl-tRNA synthetase 2, mitochondrial (putative) |
| chr11_-_109613040 | 0.25 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr14_-_54491365 | 0.25 |
ENSMUST00000128231.2
|
Dad1
|
defender against cell death 1 |
| chr19_+_3372296 | 0.24 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr11_-_3864664 | 0.24 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr14_-_70391260 | 0.24 |
ENSMUST00000035612.7
|
Ccar2
|
cell cycle activator and apoptosis regulator 2 |
| chr3_+_99048379 | 0.24 |
ENSMUST00000004343.7
|
Wars2
|
tryptophanyl tRNA synthetase 2 (mitochondrial) |
| chr9_+_44893077 | 0.23 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr2_-_18042548 | 0.23 |
ENSMUST00000066163.3
|
A930004D18Rik
|
RIKEN cDNA A930004D18 gene |
| chr1_+_52026296 | 0.22 |
ENSMUST00000168302.8
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr11_+_48977888 | 0.22 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr6_+_42885812 | 0.22 |
ENSMUST00000216408.2
|
Olfr447
|
olfactory receptor 447 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.6 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 2.0 | 6.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.9 | 5.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.5 | 8.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.0 | 3.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.9 | 6.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.8 | 5.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 2.3 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.5 | 1.6 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.5 | 2.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.4 | 5.3 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.4 | 1.7 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.4 | 6.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.4 | 4.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 1.1 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.4 | 1.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.4 | 1.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.3 | 0.9 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 | 0.9 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.3 | 1.2 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.3 | 1.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.3 | 1.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 | 2.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.2 | 1.0 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 5.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 0.9 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.2 | 0.6 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.2 | 0.6 | GO:0060464 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 0.2 | 1.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 0.8 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.2 | 0.8 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 0.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 0.5 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.2 | 0.9 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.2 | 0.7 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.2 | 0.7 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 | 0.7 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 2.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.6 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 0.5 | GO:0038091 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.2 | 1.1 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 1.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 6.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 1.0 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 1.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.6 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.1 | 1.7 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 0.1 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.1 | 0.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 1.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.3 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.1 | 0.4 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.4 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.7 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 3.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.3 | GO:0035565 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.5 | GO:0042891 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 0.1 | 1.6 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.2 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.4 | GO:0035483 | gastric motility(GO:0035482) gastric emptying(GO:0035483) negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 1.1 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 2.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0070668 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.4 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 5.0 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 1.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.8 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 2.7 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 1.0 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 1.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 1.1 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 3.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.6 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 2.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.5 | GO:0071548 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 1.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.2 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.7 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.4 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 2.4 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.8 | 10.6 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.3 | 8.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 2.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 2.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 4.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 1.0 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 1.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 3.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.6 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 4.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 3.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.8 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.9 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 1.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 7.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 5.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 1.6 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.4 | 18.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.4 | 4.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 1.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 6.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 2.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 0.9 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.3 | 2.7 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.3 | 1.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 2.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.3 | 0.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 4.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.2 | 0.9 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.2 | 8.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.5 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.2 | 0.5 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.2 | 1.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 0.6 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.2 | 0.5 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.2 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 1.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.8 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.6 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 1.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 2.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 2.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.6 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 1.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 7.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 6.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 6.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.9 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 2.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 6.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.6 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 1.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 1.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 8.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.9 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.5 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 3.2 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 3.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 8.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 5.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 10.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 1.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 3.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 5.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 2.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |