Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Bcl6b
Z-value: 0.71
Transcription factors associated with Bcl6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bcl6b
|
ENSMUSG00000000317.12 | Bcl6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl6b | mm39_v1_chr11_-_70120503_70120592 | -0.52 | 1.1e-03 | Click! |
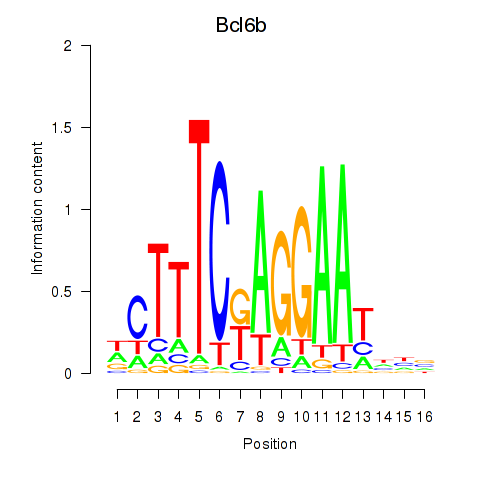
Activity profile of Bcl6b motif
Sorted Z-values of Bcl6b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Bcl6b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_46661501 | 1.36 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr10_-_86541349 | 1.31 |
ENSMUST00000020238.14
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr16_+_76810588 | 0.90 |
ENSMUST00000239066.2
ENSMUST00000023580.8 |
Usp25
|
ubiquitin specific peptidase 25 |
| chr17_-_45903494 | 0.72 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr6_+_106746081 | 0.70 |
ENSMUST00000113247.8
ENSMUST00000113249.8 ENSMUST00000113248.4 ENSMUST00000057578.16 |
Trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr17_-_45903410 | 0.65 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr13_-_98951890 | 0.64 |
ENSMUST00000040340.16
ENSMUST00000179563.8 ENSMUST00000109403.2 |
Fcho2
|
FCH domain only 2 |
| chr13_-_74956924 | 0.63 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr15_-_103123711 | 0.57 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr7_+_119499322 | 0.53 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr13_-_98951627 | 0.50 |
ENSMUST00000224992.2
ENSMUST00000225840.2 |
Fcho2
|
FCH domain only 2 |
| chr16_-_34083315 | 0.50 |
ENSMUST00000114953.8
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr16_+_93574129 | 0.49 |
ENSMUST00000228261.2
|
Dop1b
|
DOP1 leucine zipper like protein B |
| chr5_+_102629365 | 0.48 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr6_-_116050081 | 0.47 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr5_-_36771074 | 0.46 |
ENSMUST00000132383.6
ENSMUST00000174019.2 |
D5Ertd579e
Gm42936
|
DNA segment, Chr 5, ERATO Doi 579, expressed predicted gene 42936 |
| chr8_-_5155347 | 0.45 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr14_+_47535717 | 0.45 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr1_+_139382485 | 0.44 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr18_-_42084249 | 0.42 |
ENSMUST00000070949.6
ENSMUST00000235606.2 |
Prelid2
|
PRELI domain containing 2 |
| chr8_+_26091607 | 0.41 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chrX_+_73473277 | 0.41 |
ENSMUST00000114127.8
ENSMUST00000064407.10 ENSMUST00000156707.3 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr10_+_69234810 | 0.40 |
ENSMUST00000218680.2
|
Ank3
|
ankyrin 3, epithelial |
| chr5_-_150588995 | 0.40 |
ENSMUST00000118316.8
|
N4bp2l2
|
NEDD4 binding protein 2-like 2 |
| chr3_+_84832783 | 0.40 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr7_-_24245419 | 0.39 |
ENSMUST00000011776.8
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr16_-_34083549 | 0.39 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr3_+_95496239 | 0.39 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr18_+_24122689 | 0.38 |
ENSMUST00000074941.8
|
Zfp35
|
zinc finger protein 35 |
| chr7_+_48438751 | 0.37 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr2_-_77776675 | 0.37 |
ENSMUST00000111821.9
ENSMUST00000111818.8 |
Cwc22
|
CWC22 spliceosome-associated protein |
| chr3_+_95496270 | 0.37 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr6_+_34575435 | 0.37 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr4_+_3574855 | 0.36 |
ENSMUST00000052712.6
|
Tgs1
|
trimethylguanosine synthase 1 |
| chr7_+_99659121 | 0.33 |
ENSMUST00000107084.8
|
Chrdl2
|
chordin-like 2 |
| chr16_-_34083200 | 0.31 |
ENSMUST00000114947.2
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr18_-_37777238 | 0.31 |
ENSMUST00000066272.6
|
Taf7
|
TATA-box binding protein associated factor 7 |
| chr15_-_12549963 | 0.31 |
ENSMUST00000189324.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr3_+_103821413 | 0.30 |
ENSMUST00000051139.13
ENSMUST00000068879.11 |
Rsbn1
|
rosbin, round spermatid basic protein 1 |
| chr11_-_73290321 | 0.30 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr6_+_149210941 | 0.30 |
ENSMUST00000190785.7
ENSMUST00000189932.7 ENSMUST00000100765.11 |
Resf1
|
retroelement silencing factor 1 |
| chr6_+_149210889 | 0.29 |
ENSMUST00000130664.8
ENSMUST00000046689.13 |
Resf1
|
retroelement silencing factor 1 |
| chr5_+_102629240 | 0.29 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr3_-_103553347 | 0.29 |
ENSMUST00000117271.3
|
Atg4a-ps
|
autophagy related 4A, pseudogene |
| chr2_+_130975417 | 0.28 |
ENSMUST00000110225.2
|
Gm11037
|
predicted gene 11037 |
| chr18_+_57666852 | 0.28 |
ENSMUST00000079738.10
ENSMUST00000135806.8 ENSMUST00000127130.9 |
Ccdc192
|
coiled-coil domain containing 192 |
| chr2_-_77776719 | 0.27 |
ENSMUST00000065889.10
|
Cwc22
|
CWC22 spliceosome-associated protein |
| chr3_+_55149947 | 0.27 |
ENSMUST00000167204.8
ENSMUST00000054237.14 |
Dclk1
|
doublecortin-like kinase 1 |
| chr4_+_155646807 | 0.26 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chr6_+_48963795 | 0.26 |
ENSMUST00000037696.6
|
Svs1
|
seminal vesicle secretory protein 1 |
| chr16_+_48877762 | 0.25 |
ENSMUST00000168680.2
|
Myh15
|
myosin, heavy chain 15 |
| chr9_-_109531768 | 0.25 |
ENSMUST00000098359.4
|
Fbxw18
|
F-box and WD-40 domain protein 18 |
| chr10_-_115198093 | 0.25 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr12_-_40184174 | 0.24 |
ENSMUST00000078481.14
ENSMUST00000002640.6 |
Scin
|
scinderin |
| chr16_-_74208180 | 0.24 |
ENSMUST00000117200.8
|
Robo2
|
roundabout guidance receptor 2 |
| chr3_-_144412394 | 0.24 |
ENSMUST00000200532.2
|
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr3_+_144283355 | 0.24 |
ENSMUST00000151086.3
|
Selenof
|
selenoprotein F |
| chr12_+_104280812 | 0.24 |
ENSMUST00000109957.3
|
Serpina3j
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3J |
| chr8_+_96260694 | 0.23 |
ENSMUST00000041569.5
|
Ccdc113
|
coiled-coil domain containing 113 |
| chr6_-_129303659 | 0.23 |
ENSMUST00000203159.2
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr7_+_17546558 | 0.23 |
ENSMUST00000023953.5
|
Ceacam14
|
carcinoembryonic antigen-related cell adhesion molecule 14 |
| chr5_-_92511136 | 0.22 |
ENSMUST00000077820.6
|
Cxcl11
|
chemokine (C-X-C motif) ligand 11 |
| chr5_-_150588797 | 0.22 |
ENSMUST00000141857.2
|
N4bp2l2
|
NEDD4 binding protein 2-like 2 |
| chr8_+_27532583 | 0.22 |
ENSMUST00000033875.10
ENSMUST00000209525.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr2_-_73316809 | 0.21 |
ENSMUST00000141264.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr3_+_122717852 | 0.21 |
ENSMUST00000106429.6
|
1810037I17Rik
|
RIKEN cDNA 1810037I17 gene |
| chr18_-_3280999 | 0.21 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr9_+_8544143 | 0.20 |
ENSMUST00000050433.8
ENSMUST00000217462.2 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr9_-_109025043 | 0.20 |
ENSMUST00000061456.8
|
Fbxw13
|
F-box and WD-40 domain protein 13 |
| chr6_-_57992144 | 0.20 |
ENSMUST00000228070.2
ENSMUST00000228040.2 |
Vmn1r26
|
vomeronasal 1 receptor 26 |
| chr5_-_138153956 | 0.20 |
ENSMUST00000132318.2
ENSMUST00000049393.15 |
Zfp113
|
zinc finger protein 113 |
| chr3_-_88280047 | 0.20 |
ENSMUST00000107543.8
ENSMUST00000107542.2 |
Bglap3
|
bone gamma-carboxyglutamate protein 3 |
| chr16_+_9988080 | 0.20 |
ENSMUST00000121292.8
ENSMUST00000044103.6 |
Rpl39l
|
ribosomal protein L39-like |
| chr18_-_3281089 | 0.19 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr9_-_108991088 | 0.19 |
ENSMUST00000199540.2
ENSMUST00000198076.5 ENSMUST00000054925.13 |
Fbxw21
|
F-box and WD-40 domain protein 21 |
| chr12_+_104082229 | 0.19 |
ENSMUST00000021496.8
|
Serpina3a
|
serine (or cysteine) peptidase inhibitor, clade A, member 3A |
| chr9_+_40180726 | 0.18 |
ENSMUST00000171835.9
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr4_-_96236887 | 0.18 |
ENSMUST00000015368.8
|
Cyp2j11
|
cytochrome P450, family 2, subfamily j, polypeptide 11 |
| chr17_-_73706284 | 0.18 |
ENSMUST00000095208.4
|
Capn13
|
calpain 13 |
| chr4_-_57956411 | 0.17 |
ENSMUST00000030051.6
|
Txn1
|
thioredoxin 1 |
| chr6_+_56947258 | 0.17 |
ENSMUST00000226130.2
ENSMUST00000228276.2 |
Vmn1r5
|
vomeronasal 1 receptor 5 |
| chr5_+_87814058 | 0.17 |
ENSMUST00000199506.5
ENSMUST00000197631.5 ENSMUST00000094641.9 |
Csn1s1
|
casein alpha s1 |
| chr15_-_94302139 | 0.17 |
ENSMUST00000035342.11
ENSMUST00000155907.2 |
Adamts20
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 20 |
| chr9_-_109575157 | 0.17 |
ENSMUST00000071917.4
|
Fbxw26
|
F-box and WD-40 domain protein 26 |
| chr10_-_78453908 | 0.16 |
ENSMUST00000095473.6
ENSMUST00000213877.2 ENSMUST00000203305.3 |
Olfr1357
|
olfactory receptor 1357 |
| chr11_+_95603494 | 0.16 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr17_+_23562715 | 0.16 |
ENSMUST00000168175.3
ENSMUST00000234796.2 |
Vmn2r115
|
vomeronasal 2, receptor 115 |
| chr6_+_78347844 | 0.16 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr5_-_123859070 | 0.15 |
ENSMUST00000031376.12
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr8_-_118528643 | 0.15 |
ENSMUST00000034303.3
|
Mphosph6
|
M phase phosphoprotein 6 |
| chr3_+_64884839 | 0.15 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr16_-_74208395 | 0.15 |
ENSMUST00000227347.2
|
Robo2
|
roundabout guidance receptor 2 |
| chr4_+_106954794 | 0.15 |
ENSMUST00000221740.2
|
Cdcp2
|
CUB domain containing protein 2 |
| chr7_+_43228999 | 0.14 |
ENSMUST00000058104.8
ENSMUST00000205769.2 |
Zfp719
|
zinc finger protein 719 |
| chr3_-_92528480 | 0.14 |
ENSMUST00000170676.3
|
Lce6a
|
late cornified envelope 6A |
| chr13_+_50852348 | 0.14 |
ENSMUST00000099518.4
|
Gm8765
|
predicted gene 8765 |
| chr10_-_115197775 | 0.14 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr9_-_109168710 | 0.13 |
ENSMUST00000196351.2
ENSMUST00000200156.5 ENSMUST00000112040.9 ENSMUST00000112039.7 |
Fbxw28
|
F-box and WD-40 domain protein 28 |
| chr8_+_12965876 | 0.13 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr9_-_109116739 | 0.13 |
ENSMUST00000112041.6
ENSMUST00000198844.5 |
Fbxw14
|
F-box and WD-40 domain protein 14 |
| chr6_-_57938488 | 0.13 |
ENSMUST00000228097.2
|
Vmn1r24
|
vomeronasal 1 receptor 24 |
| chr6_+_57043772 | 0.13 |
ENSMUST00000228714.2
ENSMUST00000227399.2 |
Vmn1r9
|
vomeronasal 1 receptor 9 |
| chr7_+_104210108 | 0.13 |
ENSMUST00000219111.2
ENSMUST00000215410.2 ENSMUST00000216131.2 |
Olfr652
|
olfactory receptor 652 |
| chr9_-_109397316 | 0.13 |
ENSMUST00000198112.2
ENSMUST00000198397.5 ENSMUST00000056745.12 |
Fbxw15
|
F-box and WD-40 domain protein 15 |
| chrY_-_9416148 | 0.13 |
ENSMUST00000190315.2
|
Gm21874
|
predicted gene, 21874 |
| chr5_-_35682886 | 0.12 |
ENSMUST00000132959.2
|
Cpz
|
carboxypeptidase Z |
| chr3_+_79791798 | 0.12 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr6_+_41165156 | 0.12 |
ENSMUST00000103277.2
|
Trbv20
|
T cell receptor beta, variable 20 |
| chr6_+_70332836 | 0.12 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr1_+_21310803 | 0.12 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr2_+_18703797 | 0.12 |
ENSMUST00000095132.10
|
Spag6
|
sperm associated antigen 6 |
| chr2_+_18703863 | 0.11 |
ENSMUST00000173763.2
|
Spag6
|
sperm associated antigen 6 |
| chr6_+_106746138 | 0.11 |
ENSMUST00000205163.2
|
Trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr3_-_151953894 | 0.11 |
ENSMUST00000196529.5
|
Nexn
|
nexilin |
| chr1_-_173770010 | 0.11 |
ENSMUST00000042228.15
ENSMUST00000081216.12 ENSMUST00000129829.8 ENSMUST00000123708.8 |
Ifi203
|
interferon activated gene 203 |
| chr5_-_123859153 | 0.11 |
ENSMUST00000196282.5
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr5_+_87955941 | 0.10 |
ENSMUST00000072539.12
ENSMUST00000113279.8 ENSMUST00000101057.8 ENSMUST00000197301.2 |
Csn1s2b
|
casein alpha s2-like B |
| chrX_+_162923474 | 0.10 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr11_+_58323385 | 0.10 |
ENSMUST00000108825.2
|
Gm12253
|
predicted gene 12253 |
| chr10_+_81469522 | 0.09 |
ENSMUST00000140345.2
ENSMUST00000126323.2 |
Ankrd24
|
ankyrin repeat domain 24 |
| chr1_+_21310821 | 0.09 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr5_-_87634665 | 0.09 |
ENSMUST00000201519.2
|
Gm43638
|
predicted gene 43638 |
| chr9_+_37714354 | 0.08 |
ENSMUST00000215287.2
|
Olfr876
|
olfactory receptor 876 |
| chr17_-_55932192 | 0.08 |
ENSMUST00000168440.3
|
Vmn2r118
|
vomeronasal 2, receptor 118 |
| chr6_+_18170686 | 0.08 |
ENSMUST00000045706.12
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr10_+_97442727 | 0.07 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr6_-_66595266 | 0.07 |
ENSMUST00000227418.2
|
Vmn1r33
|
vomeronasal 1 receptor 33 |
| chr1_+_88234454 | 0.07 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr6_-_66620810 | 0.06 |
ENSMUST00000228647.2
ENSMUST00000227332.2 ENSMUST00000226262.2 ENSMUST00000226999.2 |
Vmn1r34
|
vomeronasal 1 receptor 34 |
| chr5_-_87847268 | 0.06 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr3_-_37180093 | 0.06 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr7_-_45071897 | 0.06 |
ENSMUST00000210271.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr18_-_65527078 | 0.06 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr7_+_29607917 | 0.05 |
ENSMUST00000186475.2
|
Zfp383
|
zinc finger protein 383 |
| chr1_-_173707677 | 0.05 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chrX_+_150127171 | 0.05 |
ENSMUST00000073364.6
|
Fam120c
|
family with sequence similarity 120, member C |
| chr17_+_37769807 | 0.05 |
ENSMUST00000214668.2
ENSMUST00000217602.2 ENSMUST00000214938.2 |
Olfr109
|
olfactory receptor 109 |
| chr5_-_74692327 | 0.04 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr6_+_78347636 | 0.03 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr10_-_95253042 | 0.03 |
ENSMUST00000135822.8
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr19_-_40260060 | 0.03 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr1_-_37575313 | 0.03 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr5_+_20433169 | 0.03 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_-_18968979 | 0.02 |
ENSMUST00000144988.8
|
Meis1
|
Meis homeobox 1 |
| chr18_+_37453427 | 0.02 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chrX_+_163219983 | 0.02 |
ENSMUST00000036858.11
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr6_+_57086439 | 0.01 |
ENSMUST00000228270.2
|
Vmn1r10
|
vomeronasal 1 receptor 10 |
| chr18_+_44403169 | 0.01 |
ENSMUST00000042747.4
|
Npy6r
|
neuropeptide Y receptor Y6 |
| chr7_+_55443967 | 0.01 |
ENSMUST00000206454.2
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr5_+_45677571 | 0.01 |
ENSMUST00000156481.8
ENSMUST00000119579.3 ENSMUST00000118833.3 |
Med28
|
mediator complex subunit 28 |
| chr6_+_18170757 | 0.01 |
ENSMUST00000129452.8
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.2 | 1.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 1.2 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.4 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 0.2 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.1 | 1.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.2 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 1.4 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.9 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.8 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.4 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.1 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.2 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.0 | 0.4 | GO:0009452 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.3 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 1.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.0 | 1.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 1.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |