Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Brca1
Z-value: 1.44
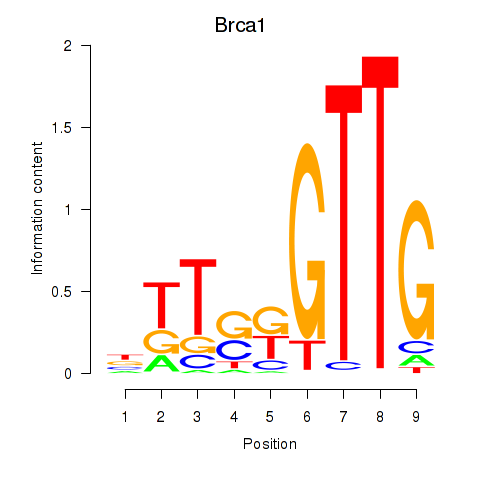
Transcription factors associated with Brca1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Brca1
|
ENSMUSG00000017146.13 | Brca1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Brca1 | mm39_v1_chr11_-_101442663_101442719 | 0.84 | 1.9e-10 | Click! |
Activity profile of Brca1 motif
Sorted Z-values of Brca1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Brca1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90638580 | 17.72 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr17_+_41121979 | 12.58 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr11_+_87684548 | 11.26 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr7_+_100143250 | 7.55 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr8_+_85696453 | 5.53 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr15_-_36609812 | 5.49 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_100396635 | 5.49 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr11_-_99328969 | 5.43 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr8_+_85696396 | 5.34 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr11_-_106205320 | 4.96 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr7_+_27147403 | 4.65 |
ENSMUST00000037399.16
ENSMUST00000108358.8 |
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr12_+_83567240 | 4.65 |
ENSMUST00000021645.9
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr9_+_96140781 | 4.65 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr1_-_171108754 | 4.60 |
ENSMUST00000073120.11
|
Ppox
|
protoporphyrinogen oxidase |
| chr19_-_10181243 | 4.56 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr2_+_164790139 | 4.55 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr6_-_125168453 | 4.17 |
ENSMUST00000189959.2
|
Ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr17_+_6697511 | 4.00 |
ENSMUST00000179569.3
|
Dynlt1b
|
dynein light chain Tctex-type 1B |
| chrX_+_134894573 | 4.00 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr13_-_100912308 | 3.92 |
ENSMUST00000075550.4
|
Cenph
|
centromere protein H |
| chr15_+_79578141 | 3.90 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr5_+_136982100 | 3.73 |
ENSMUST00000111094.8
ENSMUST00000111097.8 |
Fis1
|
fission, mitochondrial 1 |
| chr5_+_146769915 | 3.59 |
ENSMUST00000075453.9
ENSMUST00000099272.3 |
Rpl21
|
ribosomal protein L21 |
| chr6_-_122778598 | 3.51 |
ENSMUST00000165884.8
|
Slc2a3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr19_-_6191440 | 3.49 |
ENSMUST00000025893.7
|
Arl2
|
ADP-ribosylation factor-like 2 |
| chr5_+_146769700 | 3.49 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chrX_-_149372840 | 3.28 |
ENSMUST00000112725.8
ENSMUST00000112720.8 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr7_-_55669702 | 3.28 |
ENSMUST00000052204.6
|
Nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) |
| chr8_+_85598734 | 3.26 |
ENSMUST00000170296.2
ENSMUST00000136026.8 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr13_+_28441511 | 3.14 |
ENSMUST00000223428.2
|
Rps18-ps5
|
ribosomal protein S18, pseudogene 5 |
| chr4_-_133600308 | 3.13 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chrX_-_100266032 | 3.05 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr5_+_137756407 | 2.98 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr5_-_148988110 | 2.84 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr1_-_169358912 | 2.83 |
ENSMUST00000192248.2
ENSMUST00000028000.13 |
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr11_-_84710276 | 2.77 |
ENSMUST00000018549.8
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 |
| chr13_-_97897139 | 2.76 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chr9_+_96140750 | 2.73 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr5_-_73413888 | 2.58 |
ENSMUST00000101127.12
|
Fryl
|
FRY like transcription coactivator |
| chr12_-_69205882 | 2.54 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr9_+_119956755 | 2.51 |
ENSMUST00000035105.7
ENSMUST00000217317.2 |
Rpsa
|
ribosomal protein SA |
| chr18_-_43610829 | 2.43 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr19_-_32443978 | 2.41 |
ENSMUST00000078034.5
|
Rpl9-ps6
|
ribosomal protein L9, pseudogene 6 |
| chr2_-_152673585 | 2.37 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr9_+_64718708 | 2.34 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr13_+_100788087 | 2.23 |
ENSMUST00000190165.7
ENSMUST00000185767.7 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr13_+_55517545 | 2.21 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr15_-_76524097 | 2.17 |
ENSMUST00000168185.8
|
Tonsl
|
tonsoku-like, DNA repair protein |
| chr17_+_47922497 | 2.12 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr9_+_64718596 | 2.01 |
ENSMUST00000038890.6
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr13_+_100787841 | 2.01 |
ENSMUST00000084721.8
ENSMUST00000190729.2 |
Ak6
|
adenylate kinase 6 |
| chr4_+_134847949 | 1.98 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3 |
| chr2_-_155424576 | 1.92 |
ENSMUST00000126322.8
|
Gss
|
glutathione synthetase |
| chr15_+_81900570 | 1.91 |
ENSMUST00000069530.13
ENSMUST00000168581.8 ENSMUST00000164779.2 |
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr17_-_47922374 | 1.90 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr12_+_83567303 | 1.77 |
ENSMUST00000222502.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr3_-_102111892 | 1.75 |
ENSMUST00000029453.13
|
Vangl1
|
VANGL planar cell polarity 1 |
| chr19_+_53298906 | 1.72 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr19_+_10582987 | 1.66 |
ENSMUST00000237337.2
|
Ddb1
|
damage specific DNA binding protein 1 |
| chr4_-_41464816 | 1.66 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr7_+_101546059 | 1.56 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr7_+_27252658 | 1.51 |
ENSMUST00000067386.14
ENSMUST00000191126.7 ENSMUST00000187960.7 |
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr7_-_27252543 | 1.47 |
ENSMUST00000127240.8
ENSMUST00000117095.8 ENSMUST00000117611.8 |
Pld3
|
phospholipase D family, member 3 |
| chr15_+_102368510 | 1.45 |
ENSMUST00000164688.2
|
Prr13
|
proline rich 13 |
| chr17_+_36134398 | 1.42 |
ENSMUST00000173493.8
ENSMUST00000173147.8 |
Flot1
|
flotillin 1 |
| chr11_-_104441218 | 1.41 |
ENSMUST00000106962.9
ENSMUST00000106961.2 ENSMUST00000093923.9 |
Cdc27
|
cell division cycle 27 |
| chr17_-_33904345 | 1.41 |
ENSMUST00000234474.2
ENSMUST00000139302.8 ENSMUST00000114385.9 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr5_+_64969679 | 1.41 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr18_+_36926929 | 1.41 |
ENSMUST00000001419.10
|
Zmat2
|
zinc finger, matrin type 2 |
| chr6_-_142418801 | 1.39 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr6_-_120334382 | 1.36 |
ENSMUST00000032283.12
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr10_+_127512933 | 1.36 |
ENSMUST00000118612.8
ENSMUST00000048099.5 |
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr17_+_36152383 | 1.31 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr2_+_29533799 | 1.29 |
ENSMUST00000238899.2
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr7_+_101545547 | 1.25 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr17_-_33904386 | 1.24 |
ENSMUST00000087582.13
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr3_+_87837566 | 1.24 |
ENSMUST00000055984.7
|
Isg20l2
|
interferon stimulated exonuclease gene 20-like 2 |
| chr11_+_104441489 | 1.13 |
ENSMUST00000018800.9
|
Myl4
|
myosin, light polypeptide 4 |
| chr6_-_136758716 | 1.08 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr15_+_102392586 | 1.06 |
ENSMUST00000229061.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr6_-_122317484 | 1.06 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr18_-_47466378 | 1.06 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr6_-_120334400 | 1.03 |
ENSMUST00000112703.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr14_-_61283911 | 1.02 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr11_+_78717398 | 1.01 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr10_-_30531832 | 1.01 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr8_+_55024446 | 0.96 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr19_+_10181378 | 0.96 |
ENSMUST00000040372.14
|
Tmem258
|
transmembrane protein 258 |
| chr5_-_123127346 | 0.96 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr9_-_96246460 | 0.93 |
ENSMUST00000034983.7
|
Atp1b3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr7_-_46558754 | 0.92 |
ENSMUST00000209538.2
|
Tsg101
|
tumor susceptibility gene 101 |
| chrX_+_168468186 | 0.91 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr19_+_36061096 | 0.91 |
ENSMUST00000025714.9
|
Rpp30
|
ribonuclease P/MRP 30 subunit |
| chr15_+_102427149 | 0.84 |
ENSMUST00000146756.8
ENSMUST00000142194.3 |
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chr2_-_111843053 | 0.83 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr8_-_61407760 | 0.82 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr14_+_30723340 | 0.81 |
ENSMUST00000168584.9
ENSMUST00000226378.2 |
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr7_+_35148188 | 0.79 |
ENSMUST00000118383.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr10_-_30531768 | 0.77 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr19_+_44551280 | 0.75 |
ENSMUST00000040455.5
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr9_+_8544143 | 0.69 |
ENSMUST00000050433.8
ENSMUST00000217462.2 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr7_-_19449319 | 0.68 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr13_-_49473695 | 0.67 |
ENSMUST00000110086.2
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr4_-_123507494 | 0.67 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr11_-_41891111 | 0.66 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr6_-_120334302 | 0.65 |
ENSMUST00000163827.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr8_-_85696369 | 0.64 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr7_-_100581314 | 0.63 |
ENSMUST00000107032.3
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr2_-_134485936 | 0.63 |
ENSMUST00000110120.2
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr4_+_106768714 | 0.63 |
ENSMUST00000072753.13
ENSMUST00000097934.10 |
Ssbp3
|
single-stranded DNA binding protein 3 |
| chr6_-_122317457 | 0.63 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chr4_-_98705774 | 0.60 |
ENSMUST00000102790.4
|
Kank4
|
KN motif and ankyrin repeat domains 4 |
| chr17_+_71511642 | 0.58 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr13_+_93441447 | 0.55 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr4_+_106768662 | 0.55 |
ENSMUST00000030367.15
ENSMUST00000149926.8 |
Ssbp3
|
single-stranded DNA binding protein 3 |
| chr2_-_29677634 | 0.55 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr4_+_108336164 | 0.54 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr10_-_13264497 | 0.50 |
ENSMUST00000105546.8
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr6_-_68857658 | 0.49 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr10_-_127177729 | 0.48 |
ENSMUST00000026474.5
ENSMUST00000219671.2 |
Gli1
|
GLI-Kruppel family member GLI1 |
| chr2_+_167774247 | 0.48 |
ENSMUST00000029053.8
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr5_-_123804745 | 0.47 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr8_+_47070326 | 0.47 |
ENSMUST00000211115.2
ENSMUST00000093517.7 |
Casp3
|
caspase 3 |
| chrY_-_7169293 | 0.46 |
ENSMUST00000189201.2
|
Gm21244
|
predicted gene, 21244 |
| chr11_-_93776580 | 0.46 |
ENSMUST00000066888.10
|
Utp18
|
UTP18 small subunit processome component |
| chr7_-_112968533 | 0.43 |
ENSMUST00000047091.14
ENSMUST00000119278.8 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chr12_-_12990584 | 0.42 |
ENSMUST00000130990.2
|
Mycn
|
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
| chr19_-_12742811 | 0.42 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr6_-_148846247 | 0.41 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr7_-_18266150 | 0.41 |
ENSMUST00000094795.5
|
Psg25
|
pregnancy-specific glycoprotein 25 |
| chrX_+_73352694 | 0.40 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr7_-_81216687 | 0.39 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr4_+_109092459 | 0.38 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr5_+_67126304 | 0.38 |
ENSMUST00000132991.5
|
Limch1
|
LIM and calponin homology domains 1 |
| chr2_-_134486039 | 0.37 |
ENSMUST00000038228.11
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr1_+_179788675 | 0.36 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr12_+_28699598 | 0.35 |
ENSMUST00000020959.9
|
Rnaseh1
|
ribonuclease H1 |
| chr1_-_132635042 | 0.33 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chrX_+_41591476 | 0.32 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr10_+_69369854 | 0.32 |
ENSMUST00000182557.8
|
Ank3
|
ankyrin 3, epithelial |
| chr2_-_34760960 | 0.30 |
ENSMUST00000028225.12
|
Psmd5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr8_+_47070461 | 0.28 |
ENSMUST00000210534.2
|
Casp3
|
caspase 3 |
| chr1_+_187995096 | 0.27 |
ENSMUST00000060479.14
|
Ush2a
|
usherin |
| chr7_-_64522741 | 0.27 |
ENSMUST00000094331.5
|
Nsmce3
|
NSE3 homolog, SMC5-SMC6 complex component |
| chr14_-_52248324 | 0.26 |
ENSMUST00000226964.2
|
Zfp219
|
zinc finger protein 219 |
| chr3_+_109481223 | 0.26 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr7_-_29772226 | 0.25 |
ENSMUST00000183115.8
ENSMUST00000182919.8 ENSMUST00000183190.2 ENSMUST00000080834.15 |
Zfp82
|
zinc finger protein 82 |
| chr3_-_90340910 | 0.25 |
ENSMUST00000196530.2
|
Ints3
|
integrator complex subunit 3 |
| chrX_+_41591355 | 0.24 |
ENSMUST00000189753.7
|
Sh2d1a
|
SH2 domain containing 1A |
| chr19_-_57227742 | 0.24 |
ENSMUST00000111559.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr2_-_45000389 | 0.23 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_138019523 | 0.22 |
ENSMUST00000171498.3
ENSMUST00000085886.3 |
Smok3a
|
sperm motility kinase 3A |
| chr10_-_13264574 | 0.21 |
ENSMUST00000079698.7
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr16_-_16929056 | 0.21 |
ENSMUST00000232481.2
|
Ppil2
|
peptidylprolyl isomerase (cyclophilin)-like 2 |
| chr5_+_20112704 | 0.20 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_-_132635078 | 0.20 |
ENSMUST00000187861.7
|
Nfasc
|
neurofascin |
| chr7_+_102423702 | 0.18 |
ENSMUST00000098217.3
|
Olfr561
|
olfactory receptor 561 |
| chr10_-_7162196 | 0.17 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3 |
| chr3_-_61273228 | 0.17 |
ENSMUST00000066298.3
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr6_-_55109960 | 0.17 |
ENSMUST00000003568.15
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr4_+_10874537 | 0.16 |
ENSMUST00000101504.3
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chr16_+_44215136 | 0.15 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr13_+_99321241 | 0.14 |
ENSMUST00000056558.11
|
Zfp366
|
zinc finger protein 366 |
| chr12_+_108145802 | 0.14 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr3_-_59060907 | 0.13 |
ENSMUST00000196081.5
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr13_-_113317431 | 0.13 |
ENSMUST00000038212.14
|
Gzmk
|
granzyme K |
| chr6_-_70194405 | 0.13 |
ENSMUST00000103384.2
|
Igkv8-24
|
immunoglobulin kappa chain variable 8-24 |
| chr5_+_20112500 | 0.13 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_-_127048280 | 0.13 |
ENSMUST00000053392.11
|
Zfp689
|
zinc finger protein 689 |
| chr11_-_58521327 | 0.13 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr19_+_10502612 | 0.12 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr11_+_70861007 | 0.12 |
ENSMUST00000018593.10
|
Rpain
|
RPA interacting protein |
| chr5_+_67125902 | 0.10 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr11_-_41891359 | 0.10 |
ENSMUST00000070735.10
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr6_-_55109826 | 0.10 |
ENSMUST00000164012.3
ENSMUST00000212633.2 |
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr3_-_51184895 | 0.10 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr18_+_37138256 | 0.09 |
ENSMUST00000115658.6
|
Pcdha11
|
protocadherin alpha 11 |
| chr17_+_34129221 | 0.08 |
ENSMUST00000174541.9
|
Daxx
|
Fas death domain-associated protein |
| chr11_+_115671523 | 0.08 |
ENSMUST00000239299.2
|
Tmem94
|
transmembrane protein 94 |
| chrX_+_41591410 | 0.07 |
ENSMUST00000005839.11
|
Sh2d1a
|
SH2 domain containing 1A |
| chr17_-_52117894 | 0.07 |
ENSMUST00000176669.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr8_-_96580129 | 0.06 |
ENSMUST00000212628.2
ENSMUST00000040481.4 ENSMUST00000212270.2 |
Slc38a7
|
solute carrier family 38, member 7 |
| chr18_+_37085673 | 0.06 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr11_-_69712970 | 0.05 |
ENSMUST00000045771.7
|
Spem1
|
sperm maturation 1 |
| chr5_-_72800070 | 0.04 |
ENSMUST00000087213.12
|
Cnga1
|
cyclic nucleotide gated channel alpha 1 |
| chr10_+_69761314 | 0.04 |
ENSMUST00000182692.8
ENSMUST00000092433.12 |
Ank3
|
ankyrin 3, epithelial |
| chr14_+_32555912 | 0.03 |
ENSMUST00000104926.3
|
Fam170b
|
family with sequence similarity 170, member B |
| chr4_+_10874498 | 0.03 |
ENSMUST00000080517.14
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chr2_+_88217406 | 0.03 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178 |
| chr19_-_24202344 | 0.03 |
ENSMUST00000099558.5
ENSMUST00000232956.2 |
Tjp2
|
tight junction protein 2 |
| chr15_+_36174156 | 0.03 |
ENSMUST00000180159.8
ENSMUST00000057177.7 |
Polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr12_+_25024913 | 0.02 |
ENSMUST00000066652.7
ENSMUST00000220459.2 ENSMUST00000222941.2 |
Kidins220
|
kinase D-interacting substrate 220 |
| chr8_+_12965876 | 0.01 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr6_+_115578863 | 0.00 |
ENSMUST00000000449.9
|
Mkrn2
|
makorin, ring finger protein, 2 |
| chr17_-_16051295 | 0.00 |
ENSMUST00000231985.2
|
Rgmb
|
repulsive guidance molecule family member B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 3.1 | 12.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.5 | 4.6 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.0 | 17.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.0 | 3.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.9 | 2.8 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.9 | 3.7 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.9 | 2.8 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.9 | 5.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.9 | 4.6 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.9 | 10.9 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.8 | 2.5 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.8 | 5.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.7 | 3.5 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.6 | 2.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.6 | 4.7 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 4.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 1.2 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.4 | 3.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.4 | 7.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.4 | 1.9 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.3 | 3.5 | GO:0015870 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.3 | 1.7 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 0.8 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 | 3.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 0.8 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 1.0 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 0.9 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.2 | 1.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 0.8 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.2 | 3.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 0.9 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 2.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 3.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 5.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 0.5 | GO:0060032 | notochord regression(GO:0060032) |
| 0.2 | 2.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 2.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.8 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 2.8 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.8 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 0.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 3.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.4 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 3.9 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.1 | 0.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 1.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.6 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 4.0 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 0.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 3.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 2.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 2.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 2.0 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 2.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.4 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.4 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 5.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 2.7 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 1.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 2.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.1 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.6 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.2 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 8.8 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.7 | 11.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.5 | 2.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.5 | 5.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.5 | 2.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 2.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.3 | 1.9 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.3 | 3.3 | GO:0000801 | central element(GO:0000801) |
| 0.3 | 2.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 3.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 1.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 6.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 5.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 2.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 0.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 4.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 4.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 4.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 3.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 5.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 7.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.8 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.8 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 9.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 5.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.6 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 3.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 3.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 6.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 4.3 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 5.6 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 1.6 | 4.7 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 1.3 | 7.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.9 | 2.8 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.9 | 2.8 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.9 | 4.6 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.8 | 3.3 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.7 | 10.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.7 | 3.5 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.6 | 12.6 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.6 | 3.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 1.4 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 4.6 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 0.8 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.2 | 2.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 2.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.8 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.2 | 1.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 5.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 9.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 2.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 1.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 3.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 4.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 4.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.9 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 4.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 3.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 3.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 9.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 3.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 1.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 23.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 9.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 3.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 5.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 4.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 7.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 4.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.3 | 12.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 3.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 3.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 3.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 5.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 12.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 6.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 2.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 2.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 3.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 2.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |