Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
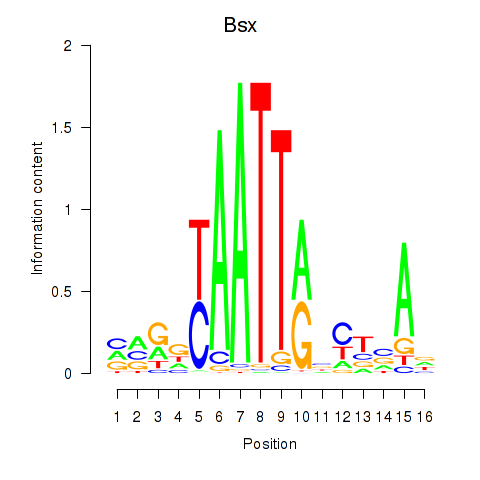
Results for Bsx
Z-value: 1.06
Transcription factors associated with Bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bsx
|
ENSMUSG00000054360.5 | Bsx |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bsx | mm39_v1_chr9_+_40785277_40785423 | 0.22 | 1.9e-01 | Click! |
Activity profile of Bsx motif
Sorted Z-values of Bsx motif
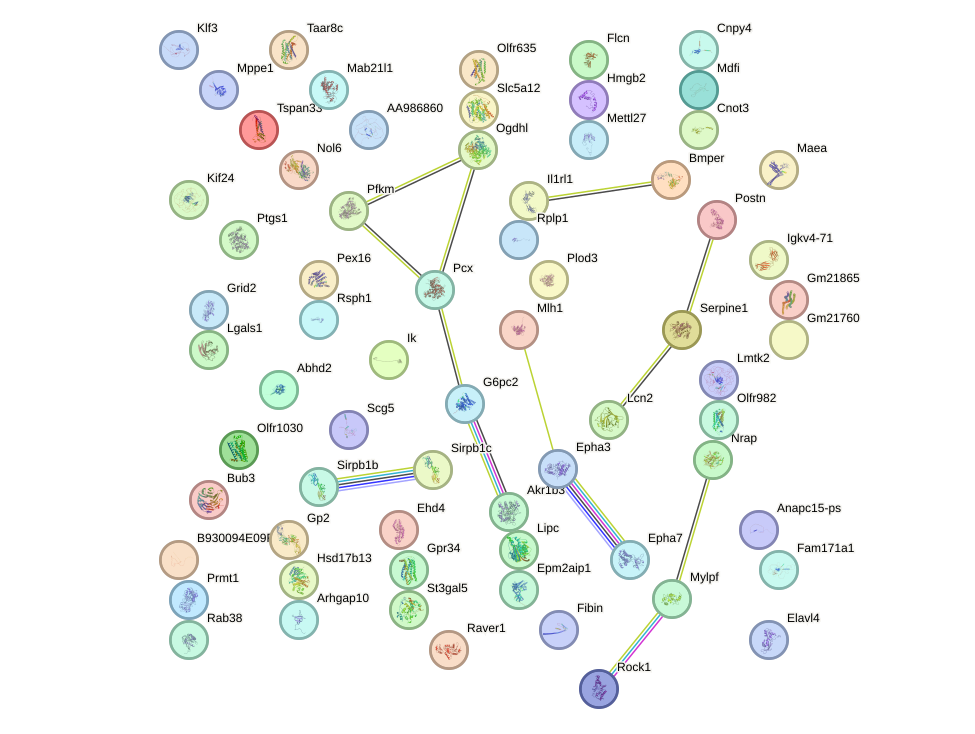
Network of associatons between targets according to the STRING database.
First level regulatory network of Bsx
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_32277773 | 4.72 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr2_-_32278245 | 4.61 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr5_-_137101108 | 3.11 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr6_+_29694181 | 3.07 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr8_+_57964921 | 2.81 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr19_+_4644425 | 2.17 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr8_+_57964956 | 2.12 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr2_-_119985078 | 2.01 |
ENSMUST00000028755.8
|
Ehd4
|
EH-domain containing 4 |
| chr15_+_78810919 | 1.95 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr7_+_126808016 | 1.87 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_-_119058489 | 1.84 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr19_+_4644365 | 1.84 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr11_+_106916430 | 1.70 |
ENSMUST00000140447.4
|
1810010H24Rik
|
RIKEN cDNA 1810010H24 gene |
| chr2_+_36120438 | 1.67 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_+_54268523 | 1.48 |
ENSMUST00000117373.8
ENSMUST00000107985.10 ENSMUST00000073012.13 ENSMUST00000081564.13 |
Postn
|
periostin, osteoblast specific factor |
| chr6_+_72074545 | 1.48 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr17_-_48144181 | 1.44 |
ENSMUST00000152455.8
ENSMUST00000035375.14 |
Mdfi
|
MyoD family inhibitor |
| chr8_-_78244412 | 1.40 |
ENSMUST00000210922.2
ENSMUST00000210519.2 |
Arhgap10
|
Rho GTPase activating protein 10 |
| chr17_-_48144094 | 1.40 |
ENSMUST00000131971.2
ENSMUST00000129360.2 ENSMUST00000113280.8 ENSMUST00000132125.8 |
Mdfi
|
MyoD family inhibitor |
| chr8_-_78244578 | 1.28 |
ENSMUST00000076316.6
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr6_-_34294377 | 1.20 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr4_-_41464816 | 1.17 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr1_+_40478787 | 1.11 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_+_130659700 | 1.05 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr2_+_3115250 | 1.03 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr7_-_44635813 | 0.98 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr9_-_61821820 | 0.93 |
ENSMUST00000008036.9
|
Rplp1
|
ribosomal protein, large, P1 |
| chr14_+_32043944 | 0.87 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr7_+_131162137 | 0.83 |
ENSMUST00000207231.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr1_+_40478926 | 0.81 |
ENSMUST00000173514.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr7_+_131162439 | 0.79 |
ENSMUST00000207442.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr5_-_138185438 | 0.77 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr7_+_88079465 | 0.76 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr2_+_85809620 | 0.75 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr7_+_131161951 | 0.74 |
ENSMUST00000084502.7
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr2_-_110193502 | 0.73 |
ENSMUST00000099626.5
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr9_-_21003268 | 0.72 |
ENSMUST00000115487.3
|
Raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr1_+_82817794 | 0.69 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr3_-_15902583 | 0.66 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr13_-_23929490 | 0.64 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr7_+_3648264 | 0.59 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr5_+_137015873 | 0.57 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr5_+_33492853 | 0.56 |
ENSMUST00000114449.7
ENSMUST00000200716.3 |
Maea
|
macrophage erythroblast attacher |
| chr7_+_101545547 | 0.53 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr5_-_138185686 | 0.51 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr9_-_70842090 | 0.50 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr13_+_21900554 | 0.47 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr6_-_50433064 | 0.45 |
ENSMUST00000146341.4
ENSMUST00000071728.11 |
Osbpl3
|
oxysterol binding protein-like 3 |
| chr7_+_131162338 | 0.43 |
ENSMUST00000208571.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr7_+_101546059 | 0.43 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr2_+_69050315 | 0.40 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr7_+_78922947 | 0.38 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr15_+_97990431 | 0.37 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr3_-_15640045 | 0.36 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr10_-_23977810 | 0.36 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr18_+_42669322 | 0.36 |
ENSMUST00000236418.2
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr18_+_31742565 | 0.35 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr1_-_173018204 | 0.33 |
ENSMUST00000215878.2
ENSMUST00000201132.3 |
Olfr1406
|
olfactory receptor 1406 |
| chr12_-_55061117 | 0.33 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr5_+_134961246 | 0.32 |
ENSMUST00000111218.8
ENSMUST00000136246.5 ENSMUST00000201847.3 |
Mettl27
|
methyltransferase like 27 |
| chr5_-_104125226 | 0.31 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr17_-_31496352 | 0.31 |
ENSMUST00000024832.9
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr5_+_64969679 | 0.29 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr5_-_104125270 | 0.29 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr10_-_6930376 | 0.28 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr18_-_67378886 | 0.27 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr14_+_32507920 | 0.27 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr2_+_85868891 | 0.26 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr6_+_56979285 | 0.25 |
ENSMUST00000079669.7
|
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr7_+_103628383 | 0.25 |
ENSMUST00000098185.2
|
Olfr635
|
olfactory receptor 635 |
| chr16_-_63684477 | 0.22 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr5_-_104125192 | 0.21 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr3_-_16060545 | 0.19 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr9_+_39985125 | 0.18 |
ENSMUST00000054051.5
|
Olfr982
|
olfactory receptor 982 |
| chrY_-_40270796 | 0.15 |
ENSMUST00000177713.2
|
Gm21865
|
predicted gene, 21865 |
| chr9_+_23285228 | 0.15 |
ENSMUST00000214050.2
|
Bmper
|
BMP-binding endothelial regulator |
| chr4_-_110143777 | 0.14 |
ENSMUST00000138972.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr5_+_134961205 | 0.14 |
ENSMUST00000047196.14
ENSMUST00000111221.9 ENSMUST00000111219.8 ENSMUST00000068617.12 |
Mettl27
|
methyltransferase like 27 |
| chr19_-_56378309 | 0.13 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr17_-_31496301 | 0.12 |
ENSMUST00000235144.2
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr2_-_88157559 | 0.11 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr3_-_16060491 | 0.10 |
ENSMUST00000108347.3
|
Gm5150
|
predicted gene 5150 |
| chr14_+_50360643 | 0.09 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr2_+_85804239 | 0.09 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr6_-_69220672 | 0.09 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr2_+_110427643 | 0.09 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr2_-_85632888 | 0.09 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr5_-_65855511 | 0.09 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chrX_+_13499008 | 0.08 |
ENSMUST00000096492.4
|
Gpr34
|
G protein-coupled receptor 34 |
| chr2_-_113659360 | 0.07 |
ENSMUST00000024005.8
|
Scg5
|
secretogranin V |
| chr3_+_55689921 | 0.07 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr11_-_59700820 | 0.06 |
ENSMUST00000047706.3
ENSMUST00000102697.10 |
Flcn
|
folliculin |
| chr4_+_28813125 | 0.06 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr9_-_70841881 | 0.06 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr5_+_138185747 | 0.06 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr18_+_36877709 | 0.05 |
ENSMUST00000007042.6
ENSMUST00000237095.2 |
Ik
|
IK cytokine |
| chr8_-_79547707 | 0.05 |
ENSMUST00000130325.8
ENSMUST00000051867.7 |
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_+_111100893 | 0.05 |
ENSMUST00000135807.2
ENSMUST00000060711.8 |
Epm2aip1
|
EPM2A (laforin) interacting protein 1 |
| chr9_-_111086528 | 0.04 |
ENSMUST00000199404.2
|
Mlh1
|
mutL homolog 1 |
| chr5_+_144037171 | 0.03 |
ENSMUST00000041804.8
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr3_-_58729732 | 0.03 |
ENSMUST00000191233.4
|
Mindy4b-ps
|
MINDY lysine 48 deubiquitinase 4B, pseudogene |
| chr4_+_28813152 | 0.02 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chr6_+_63232955 | 0.02 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr16_+_3648742 | 0.02 |
ENSMUST00000214238.2
ENSMUST00000214590.2 |
Olfr15
|
olfactory receptor 15 |
| chrY_+_80146479 | 0.02 |
ENSMUST00000179811.2
|
Gm21760
|
predicted gene, 21760 |
| chr14_+_65504067 | 0.02 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr11_-_33942981 | 0.02 |
ENSMUST00000238903.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr18_-_10182007 | 0.01 |
ENSMUST00000067947.7
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr2_+_92205651 | 0.00 |
ENSMUST00000028650.9
|
Pex16
|
peroxisomal biogenesis factor 16 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.0 | 3.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.8 | 4.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.7 | 2.0 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.5 | 4.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.4 | 1.7 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.4 | 1.2 | GO:0018931 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.4 | 1.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.4 | 1.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 1.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 0.6 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.2 | 1.9 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 0.6 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.6 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.1 | 0.8 | GO:1903232 | platelet dense granule organization(GO:0060155) phagosome acidification(GO:0090383) melanosome assembly(GO:1903232) |
| 0.1 | 2.8 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.4 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 3.7 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 2.0 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.4 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:1901874 | negative regulation of cellular respiration(GO:1901856) regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.5 | 3.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 3.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 1.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 4.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.6 | 1.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.5 | 1.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.5 | 4.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.5 | 2.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 0.8 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.2 | 1.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 1.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.6 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.1 | 0.4 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 1.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 12.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 2.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.8 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 3.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.3 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 1.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |