Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Cdx2
Z-value: 1.52
Transcription factors associated with Cdx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
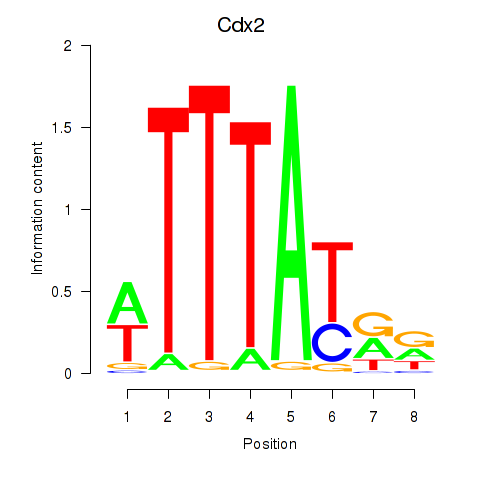
Cdx2
|
ENSMUSG00000029646.4 | Cdx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cdx2 | mm39_v1_chr5_-_147244074_147244091 | -0.23 | 1.7e-01 | Click! |
Activity profile of Cdx2 motif
Sorted Z-values of Cdx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Cdx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_90603013 | 9.86 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr1_-_45542442 | 6.72 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr7_+_130633776 | 6.18 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr8_-_106660470 | 5.71 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr11_-_99328969 | 5.62 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr14_+_80237691 | 4.82 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr3_-_14843512 | 4.06 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr17_-_31348576 | 4.00 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr6_+_38895902 | 2.92 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr4_-_63965161 | 2.89 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr6_-_49191891 | 2.86 |
ENSMUST00000031838.9
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr3_+_57332735 | 2.73 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr7_+_140343652 | 2.58 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr3_-_10273628 | 2.47 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr9_+_95519654 | 2.37 |
ENSMUST00000015498.9
|
Pcolce2
|
procollagen C-endopeptidase enhancer 2 |
| chr9_-_65330231 | 2.31 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chrX_+_162873183 | 2.22 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chrX_+_95498965 | 1.69 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr1_+_139429430 | 1.52 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr17_-_71305003 | 1.44 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr10_+_97400990 | 1.32 |
ENSMUST00000038160.6
|
Lum
|
lumican |
| chr10_+_119655294 | 1.26 |
ENSMUST00000105262.9
ENSMUST00000147454.8 ENSMUST00000138410.8 ENSMUST00000144825.8 ENSMUST00000148954.8 ENSMUST00000144959.8 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr8_-_34333573 | 1.00 |
ENSMUST00000183062.2
|
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr7_+_89814713 | 0.97 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr10_-_35587888 | 0.97 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr6_+_71176811 | 0.91 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr14_-_54923517 | 0.86 |
ENSMUST00000125265.2
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr12_-_102844537 | 0.79 |
ENSMUST00000045652.8
ENSMUST00000223554.2 |
Btbd7
|
BTB (POZ) domain containing 7 |
| chr11_-_99134885 | 0.69 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chrX_+_72818003 | 0.69 |
ENSMUST00000002081.6
|
Srpk3
|
serine/arginine-rich protein specific kinase 3 |
| chr3_-_92393193 | 0.66 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr6_-_122317484 | 0.59 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr5_-_135773047 | 0.59 |
ENSMUST00000153399.2
|
Tmem120a
|
transmembrane protein 120A |
| chr11_-_76386190 | 0.54 |
ENSMUST00000108408.9
|
Abr
|
active BCR-related gene |
| chr2_-_51039112 | 0.52 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chrX_+_162923474 | 0.42 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr11_+_96214078 | 0.36 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr4_-_9643636 | 0.33 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr17_+_80614795 | 0.31 |
ENSMUST00000223878.2
ENSMUST00000068175.6 ENSMUST00000224391.2 |
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr19_-_47680528 | 0.30 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr6_+_17491231 | 0.28 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr18_+_65715156 | 0.25 |
ENSMUST00000237553.2
ENSMUST00000237712.2 ENSMUST00000182684.8 |
Zfp532
|
zinc finger protein 532 |
| chr15_-_101389384 | 0.22 |
ENSMUST00000023718.9
|
Krt83
|
keratin 83 |
| chrM_+_7758 | 0.22 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr17_-_71153283 | 0.21 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr8_+_95113066 | 0.21 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chrM_+_7779 | 0.17 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr18_+_65715460 | 0.16 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chr19_-_34856853 | 0.14 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr14_+_54923655 | 0.11 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr5_+_13448833 | 0.02 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_+_13499008 | 0.01 |
ENSMUST00000096492.4
|
Gpr34
|
G protein-coupled receptor 34 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.7 | 6.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 2.9 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.3 | 1.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 6.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 5.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 2.3 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 1.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 2.9 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 2.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.4 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 2.6 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 4.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 4.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 1.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 0.9 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 1.0 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 1.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 4.0 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 2.9 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 3.0 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 6.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 4.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 1.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 4.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 13.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.6 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.9 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.4 | 4.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 2.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.0 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 1.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 6.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 5.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.3 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 2.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 2.9 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 2.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 2.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 5.7 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.4 | GO:0035254 | glutamate receptor binding(GO:0035254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 7.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 4.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 7.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 9.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.1 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |