Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
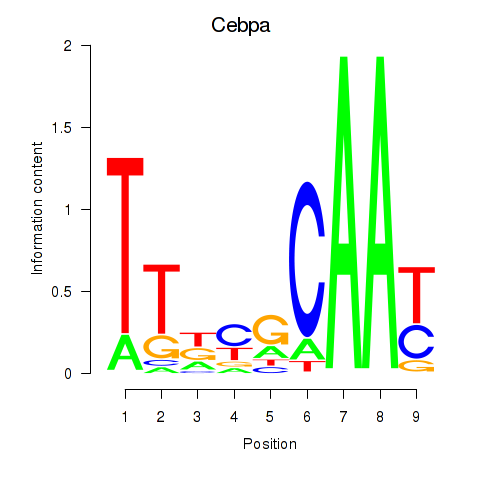
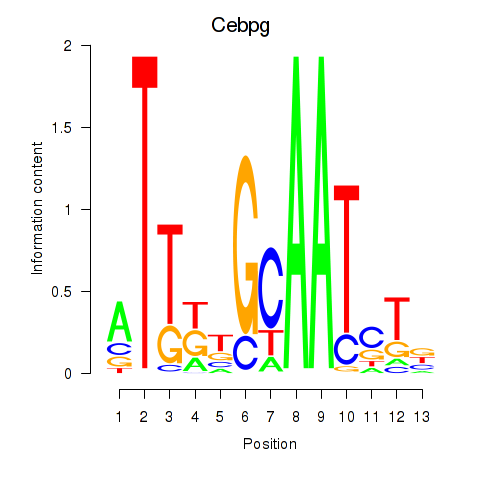
Results for Cebpa_Cebpg
Z-value: 1.66
Transcription factors associated with Cebpa_Cebpg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cebpa
|
ENSMUSG00000034957.11 | Cebpa |
|
Cebpg
|
ENSMUSG00000056216.10 | Cebpg |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpa | mm39_v1_chr7_+_34818709_34818728 | 0.70 | 1.8e-06 | Click! |
| Cebpg | mm39_v1_chr7_-_34755985_34756002 | 0.02 | 9.0e-01 | Click! |
Activity profile of Cebpa_Cebpg motif
Sorted Z-values of Cebpa_Cebpg motif
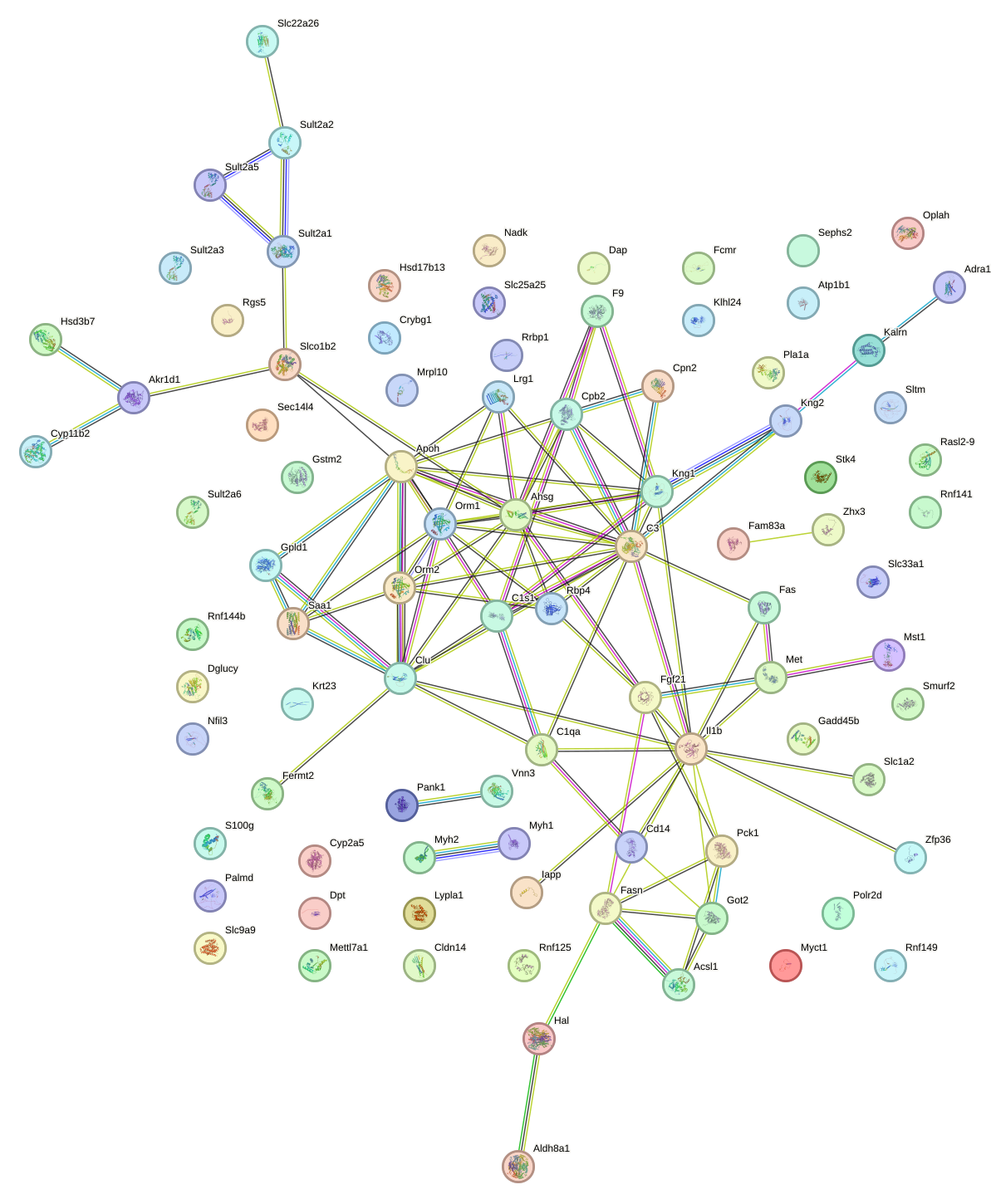
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpa_Cebpg
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13467422 | 17.39 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr7_+_13357892 | 11.38 |
ENSMUST00000108525.4
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr2_+_172994841 | 10.87 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr3_+_20011251 | 10.63 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr3_+_20011201 | 10.62 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr3_+_20011405 | 10.07 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr17_-_56428968 | 8.58 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr10_+_23727325 | 7.69 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr16_+_22710785 | 7.27 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr14_+_66208059 | 6.70 |
ENSMUST00000127387.8
|
Clu
|
clusterin |
| chr12_-_81014849 | 6.52 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr7_-_13571334 | 6.27 |
ENSMUST00000108522.5
|
Sult2a1
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr14_+_66208498 | 6.20 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr12_-_81014755 | 6.19 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr16_+_22877000 | 5.58 |
ENSMUST00000039492.14
ENSMUST00000023589.15 ENSMUST00000089902.8 |
Kng1
|
kininogen 1 |
| chr14_+_66208613 | 5.52 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr14_+_66208253 | 4.68 |
ENSMUST00000138191.8
|
Clu
|
clusterin |
| chr16_-_22847760 | 4.63 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr16_-_22847808 | 4.61 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr16_-_22847829 | 4.46 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr16_-_22848153 | 3.82 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr2_-_32314017 | 3.76 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr6_+_37507108 | 3.72 |
ENSMUST00000040987.11
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr5_+_90708962 | 3.49 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr7_+_46401214 | 3.38 |
ENSMUST00000210769.2
ENSMUST00000210272.2 ENSMUST00000075982.4 |
Saa2
|
serum amyloid A 2 |
| chr17_-_57535003 | 3.36 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr6_+_141575226 | 3.36 |
ENSMUST00000042812.9
|
Slco1b2
|
solute carrier organic anion transporter family, member 1b2 |
| chr16_-_93726399 | 3.29 |
ENSMUST00000177648.8
ENSMUST00000142083.2 |
Cldn14
|
claudin 14 |
| chr7_-_13988795 | 3.27 |
ENSMUST00000184731.8
ENSMUST00000076576.7 |
Sult2a6
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 6 |
| chr19_-_7779943 | 3.20 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr11_+_3981769 | 3.10 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr11_-_99383938 | 3.00 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr7_+_127399776 | 2.91 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr18_+_21077627 | 2.59 |
ENSMUST00000050004.3
|
Rnf125
|
ring finger protein 125 |
| chr15_-_76191301 | 2.47 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr6_-_124519240 | 2.39 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr14_-_45715308 | 2.32 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr1_+_169483062 | 2.27 |
ENSMUST00000027997.9
ENSMUST00000152809.3 |
Rgs5
|
regulator of G-protein signaling 5 |
| chr14_+_75479727 | 2.19 |
ENSMUST00000022576.10
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr7_+_26534730 | 2.13 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr19_-_7780025 | 2.12 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr7_-_46392403 | 2.12 |
ENSMUST00000128088.4
|
Saa1
|
serum amyloid A 1 |
| chr5_-_104125192 | 1.95 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_104125226 | 1.95 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_104125270 | 1.93 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr10_+_93324624 | 1.85 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase |
| chr19_+_34268053 | 1.84 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chrX_+_59044796 | 1.82 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr7_-_13856967 | 1.78 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr3_-_107893676 | 1.75 |
ENSMUST00000066530.7
ENSMUST00000012348.9 |
Gstm2
|
glutathione S-transferase, mu 2 |
| chr16_-_30086317 | 1.74 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr11_+_108286114 | 1.72 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr2_+_102536701 | 1.71 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr1_+_130793406 | 1.69 |
ENSMUST00000038829.7
|
Fcmr
|
Fc fragment of IgM receptor |
| chr13_+_47347301 | 1.67 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr19_+_34268071 | 1.64 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr10_+_21253190 | 1.62 |
ENSMUST00000042699.14
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr7_-_45264908 | 1.54 |
ENSMUST00000033099.6
|
Fgf21
|
fibroblast growth factor 21 |
| chr16_-_38253507 | 1.53 |
ENSMUST00000002926.8
|
Pla1a
|
phospholipase A1 member A |
| chr7_-_126873219 | 1.52 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr13_+_25127127 | 1.47 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr9_+_94551929 | 1.46 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr12_+_100745333 | 1.38 |
ENSMUST00000110073.8
ENSMUST00000110069.8 |
Dglucy
|
D-glutamate cyclase |
| chr9_+_107957621 | 1.32 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr3_-_116762476 | 1.32 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr9_+_107957640 | 1.28 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr15_+_100202642 | 1.27 |
ENSMUST00000067752.5
ENSMUST00000229588.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr6_+_17463748 | 1.25 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr1_+_164624200 | 1.21 |
ENSMUST00000027861.6
|
Dpt
|
dermatopontin |
| chr8_+_46924074 | 1.19 |
ENSMUST00000034046.13
ENSMUST00000211644.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr19_-_34855278 | 1.18 |
ENSMUST00000112460.3
|
Pank1
|
pantothenate kinase 1 |
| chr19_-_38113696 | 1.17 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr12_+_100745314 | 1.16 |
ENSMUST00000069782.11
|
Dglucy
|
D-glutamate cyclase |
| chr10_-_44024843 | 1.12 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr4_+_155646807 | 1.11 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chr4_+_63262775 | 1.08 |
ENSMUST00000030044.3
|
Orm1
|
orosomucoid 1 |
| chr1_-_39616445 | 0.98 |
ENSMUST00000062525.11
|
Rnf149
|
ring finger protein 149 |
| chr13_-_53135064 | 0.95 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr19_-_34855242 | 0.95 |
ENSMUST00000238065.2
|
Pank1
|
pantothenate kinase 1 |
| chr8_-_41507808 | 0.88 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr15_+_31225302 | 0.88 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr15_+_31224460 | 0.88 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr1_-_39616369 | 0.86 |
ENSMUST00000195705.2
|
Rnf149
|
ring finger protein 149 |
| chr16_+_19916292 | 0.84 |
ENSMUST00000023509.5
ENSMUST00000232088.2 ENSMUST00000231842.2 |
Klhl24
|
kelch-like 24 |
| chr15_+_31224616 | 0.83 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr1_-_164281344 | 0.83 |
ENSMUST00000193367.2
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr11_+_67090878 | 0.80 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr4_+_63280675 | 0.79 |
ENSMUST00000075341.4
|
Orm2
|
orosomucoid 2 |
| chr11_-_95966477 | 0.71 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chrX_-_161747552 | 0.69 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr11_-_106811185 | 0.68 |
ENSMUST00000167787.2
ENSMUST00000092517.9 |
Smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr11_+_96932379 | 0.66 |
ENSMUST00000001485.10
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr15_+_57849269 | 0.66 |
ENSMUST00000050374.3
|
Fam83a
|
family with sequence similarity 83, member A |
| chr11_-_43792013 | 0.66 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr2_-_160714904 | 0.63 |
ENSMUST00000109460.8
ENSMUST00000127201.2 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr10_+_80765900 | 0.60 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr15_+_31224555 | 0.59 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr2_-_160714749 | 0.58 |
ENSMUST00000176141.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr8_-_96615138 | 0.55 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr7_-_5128936 | 0.55 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr2_-_143853122 | 0.52 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr18_-_36859732 | 0.51 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr16_-_33916354 | 0.50 |
ENSMUST00000114973.9
ENSMUST00000232157.2 ENSMUST00000114964.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr3_-_63872079 | 0.50 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr3_-_63872189 | 0.47 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr7_-_13723513 | 0.47 |
ENSMUST00000165167.8
ENSMUST00000108520.4 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr15_+_102012782 | 0.46 |
ENSMUST00000230474.2
|
Tns2
|
tensin 2 |
| chr4_-_136626073 | 0.42 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr7_-_28078671 | 0.42 |
ENSMUST00000209061.2
|
Zfp36
|
zinc finger protein 36 |
| chr10_+_5543769 | 0.42 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr2_-_129213050 | 0.40 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr10_-_102326286 | 0.37 |
ENSMUST00000020040.5
|
Nts
|
neurotensin |
| chr13_+_51254852 | 0.37 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr17_+_28910393 | 0.35 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr5_-_92231517 | 0.34 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr5_+_102872838 | 0.34 |
ENSMUST00000112853.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_-_164585102 | 0.31 |
ENSMUST00000103096.10
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr7_+_51530060 | 0.31 |
ENSMUST00000145049.2
|
Gas2
|
growth arrest specific 2 |
| chr2_-_104324035 | 0.30 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chrX_+_21581135 | 0.30 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr15_-_55770118 | 0.29 |
ENSMUST00000110200.3
|
Sntb1
|
syntrophin, basic 1 |
| chr3_-_65300000 | 0.27 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chrM_+_8603 | 0.27 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr4_-_132459762 | 0.26 |
ENSMUST00000045550.5
|
Xkr8
|
X-linked Kx blood group related 8 |
| chr12_+_98234884 | 0.26 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr15_+_41615084 | 0.25 |
ENSMUST00000229511.2
ENSMUST00000229836.2 |
Oxr1
|
oxidation resistance 1 |
| chr11_+_77928736 | 0.25 |
ENSMUST00000072289.12
ENSMUST00000100784.9 ENSMUST00000073660.7 |
Flot2
|
flotillin 2 |
| chr14_+_44340111 | 0.23 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr5_-_151051000 | 0.23 |
ENSMUST00000202111.4
|
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr8_+_71823951 | 0.22 |
ENSMUST00000002469.9
ENSMUST00000110052.2 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr5_-_147831610 | 0.22 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr2_-_65397850 | 0.21 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr5_-_137856280 | 0.20 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr5_-_88823989 | 0.20 |
ENSMUST00000078945.12
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr15_+_44482667 | 0.20 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr14_-_44161016 | 0.19 |
ENSMUST00000159175.2
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr7_-_16550656 | 0.18 |
ENSMUST00000061390.9
|
Fkrp
|
fukutin related protein |
| chr12_-_113542610 | 0.18 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr16_+_3665064 | 0.17 |
ENSMUST00000041778.8
|
Zfp174
|
zinc finger protein 174 |
| chr6_-_3968365 | 0.16 |
ENSMUST00000031674.11
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr17_+_29077385 | 0.16 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr17_+_28910302 | 0.16 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr8_-_34419826 | 0.15 |
ENSMUST00000033995.14
ENSMUST00000033994.15 ENSMUST00000191473.7 ENSMUST00000053251.12 |
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr11_+_59197746 | 0.13 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chrX_+_7630182 | 0.13 |
ENSMUST00000144900.2
ENSMUST00000115677.8 ENSMUST00000101695.9 ENSMUST00000115678.3 |
Tfe3
|
transcription factor E3 |
| chr3_-_94490023 | 0.13 |
ENSMUST00000029783.16
|
Snx27
|
sorting nexin family member 27 |
| chr15_+_44482944 | 0.13 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chrX_-_93585668 | 0.12 |
ENSMUST00000026142.8
|
Maged1
|
MAGE family member D1 |
| chr6_+_30401864 | 0.12 |
ENSMUST00000068240.13
ENSMUST00000068259.10 ENSMUST00000132581.8 |
Klhdc10
|
kelch domain containing 10 |
| chr14_-_78866714 | 0.12 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr9_-_40232202 | 0.11 |
ENSMUST00000239052.2
|
Gramd1b
|
GRAM domain containing 1B |
| chr18_+_37818263 | 0.10 |
ENSMUST00000194418.2
|
Pcdhga4
|
protocadherin gamma subfamily A, 4 |
| chr2_-_65397809 | 0.09 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr2_-_52566583 | 0.08 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr4_+_150322151 | 0.08 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chr3_-_65299967 | 0.08 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr14_-_51493569 | 0.07 |
ENSMUST00000061936.8
|
Rnase2a
|
ribonuclease, RNase A family, 2A (liver, eosinophil-derived neurotoxin) |
| chr4_+_130001349 | 0.06 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr6_-_8180174 | 0.06 |
ENSMUST00000213284.2
|
Col28a1
|
collagen, type XXVIII, alpha 1 |
| chr13_-_23945189 | 0.05 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr9_+_21746785 | 0.05 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr18_-_33346885 | 0.05 |
ENSMUST00000025236.9
|
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr19_-_36097233 | 0.04 |
ENSMUST00000025718.10
|
Ankrd1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr10_+_3784877 | 0.04 |
ENSMUST00000239159.2
|
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_58625350 | 0.03 |
ENSMUST00000038372.14
ENSMUST00000097724.10 |
Fam126b
|
family with sequence similarity 126, member B |
| chrX_-_103244728 | 0.03 |
ENSMUST00000056502.7
|
Nexmif
|
neurite extension and migration factor |
| chr18_+_69633741 | 0.03 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr11_+_22970464 | 0.03 |
ENSMUST00000094363.4
ENSMUST00000151877.2 |
Fam161a
|
family with sequence similarity 161, member A |
| chr1_+_151446893 | 0.02 |
ENSMUST00000134499.8
|
Niban1
|
niban apoptosis regulator 1 |
| chr3_+_105811712 | 0.02 |
ENSMUST00000000574.3
|
Adora3
|
adenosine A3 receptor |
| chr4_-_15149051 | 0.01 |
ENSMUST00000041606.14
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr11_-_105347500 | 0.01 |
ENSMUST00000049995.10
ENSMUST00000100332.4 |
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr10_-_85847697 | 0.01 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 23.1 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.7 | 10.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.2 | 3.7 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 1.2 | 3.5 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.8 | 31.3 | GO:0006825 | copper ion transport(GO:0006825) response to copper ion(GO:0046688) |
| 0.8 | 3.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.7 | 2.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.5 | 16.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 1.5 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.4 | 1.9 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.4 | 5.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 1.8 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.3 | 19.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.3 | 2.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 2.6 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.3 | 0.8 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.3 | 0.6 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.3 | 6.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.2 | 7.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 1.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 1.5 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 0.7 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.2 | 1.5 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.2 | 0.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 8.6 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 7.7 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.2 | 1.6 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.5 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 1.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.4 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.5 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 4.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 5.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 2.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 1.2 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.5 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.5 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 2.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 3.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 2.0 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 15.7 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 5.8 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.2 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 2.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.5 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 3.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 1.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.7 | GO:0090662 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 1.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 1.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 23.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.5 | 31.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 3.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.3 | 7.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 33.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 7.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 6.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 13.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.1 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 2.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 3.3 | 23.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 2.8 | 31.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.6 | 12.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.2 | 7.3 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.9 | 23.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.9 | 3.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.7 | 3.7 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.7 | 2.9 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.5 | 1.5 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.4 | 1.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.4 | 3.4 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.4 | 2.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 3.8 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 1.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 5.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 1.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 1.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 2.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 17.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.3 | 1.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.3 | 1.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 1.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 3.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 0.6 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 0.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 1.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 1.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 15.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 9.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 4.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 2.5 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.1 | 4.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 7.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 2.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 6.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 31.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 3.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 22.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 10.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 7.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 6.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 7.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 12.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.7 | 31.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.7 | 10.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.5 | 6.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 6.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 7.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 22.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 2.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |