Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Clock
Z-value: 0.63
Transcription factors associated with Clock
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Clock
|
ENSMUSG00000029238.12 | Clock |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Clock | mm39_v1_chr5_-_76452365_76452398 | 0.32 | 5.6e-02 | Click! |
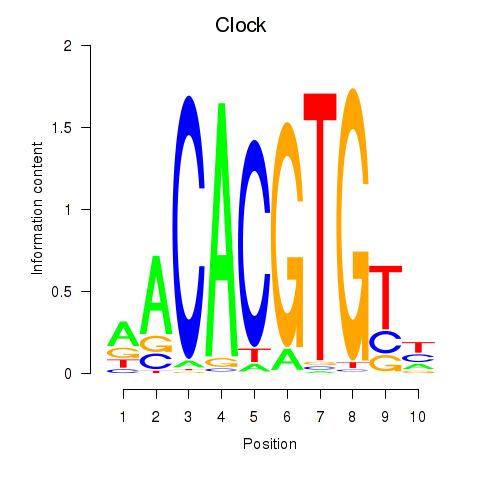
Activity profile of Clock motif
Sorted Z-values of Clock motif
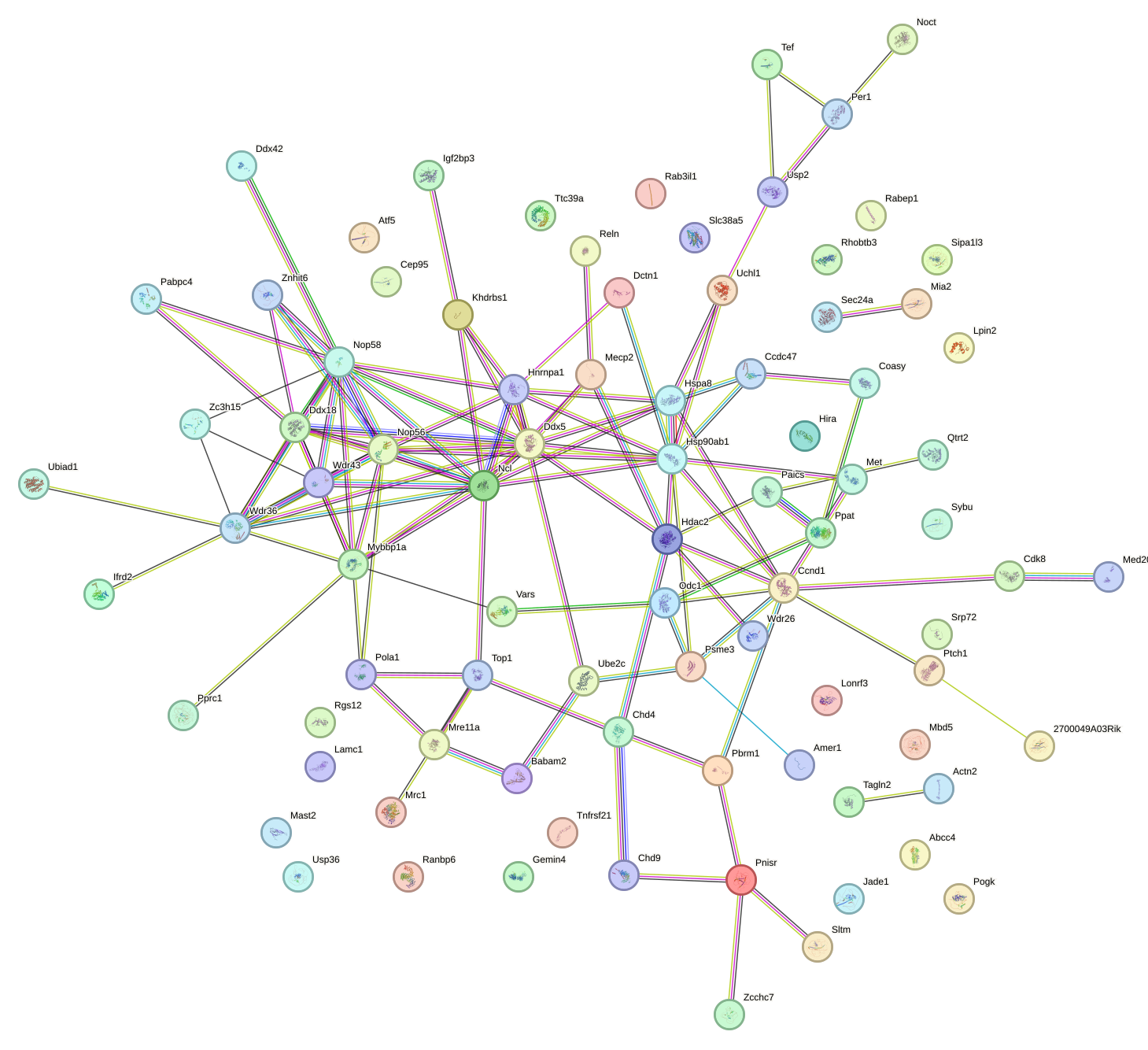
Network of associatons between targets according to the STRING database.
First level regulatory network of Clock
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_22549688 | 1.98 |
ENSMUST00000062372.14
ENSMUST00000161356.8 |
Reln
|
reelin |
| chrX_+_8137372 | 1.98 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr1_+_59724108 | 1.90 |
ENSMUST00000027174.10
ENSMUST00000190231.7 ENSMUST00000191142.7 ENSMUST00000185772.7 |
Nop58
|
NOP58 ribonucleoprotein |
| chr2_+_164611812 | 1.79 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chrX_+_8137620 | 1.75 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr15_+_103148824 | 1.71 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr2_+_160487801 | 1.68 |
ENSMUST00000109468.3
|
Top1
|
topoisomerase (DNA) I |
| chrX_+_8137881 | 1.66 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr4_+_123176570 | 1.64 |
ENSMUST00000106243.8
ENSMUST00000106241.8 ENSMUST00000080178.13 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr18_+_35686424 | 1.57 |
ENSMUST00000235679.2
ENSMUST00000235176.2 ENSMUST00000235801.2 ENSMUST00000237592.2 ENSMUST00000237230.2 ENSMUST00000237589.2 |
Snhg4
Snhg4
|
small nucleolar RNA host gene 4 small nucleolar RNA host gene 4 |
| chr3_+_51131868 | 1.44 |
ENSMUST00000023849.15
ENSMUST00000167780.2 |
Noct
|
nocturnin |
| chr4_-_131802561 | 1.25 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr4_-_131802606 | 1.17 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr11_+_68986043 | 1.16 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr18_+_42644552 | 1.13 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr11_+_101207743 | 1.12 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr6_-_49191891 | 1.11 |
ENSMUST00000031838.9
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr1_+_172327812 | 1.02 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr1_-_86286690 | 0.97 |
ENSMUST00000185785.2
|
Ncl
|
nucleolin |
| chr11_-_106679983 | 0.96 |
ENSMUST00000129585.8
|
Ddx5
|
DEAD box helicase 5 |
| chr17_+_43327412 | 0.96 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr1_-_153208477 | 0.94 |
ENSMUST00000027752.15
|
Lamc1
|
laminin, gamma 1 |
| chr5_-_77099406 | 0.94 |
ENSMUST00000140076.2
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr5_+_146168020 | 0.93 |
ENSMUST00000161181.8
ENSMUST00000161652.8 ENSMUST00000031640.15 ENSMUST00000159467.2 |
Cdk8
|
cyclin-dependent kinase 8 |
| chr13_-_12355604 | 0.92 |
ENSMUST00000168193.8
ENSMUST00000064204.14 |
Actn2
|
actinin alpha 2 |
| chr1_+_172328768 | 0.91 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr19_+_9995557 | 0.90 |
ENSMUST00000113161.10
ENSMUST00000238672.2 ENSMUST00000117641.8 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr17_+_35220252 | 0.88 |
ENSMUST00000174260.8
|
Vars
|
valyl-tRNA synthetase |
| chr11_+_72332167 | 0.86 |
ENSMUST00000045633.6
|
Mybbp1a
|
MYB binding protein (P160) 1a |
| chr11_-_106679671 | 0.82 |
ENSMUST00000123339.2
|
Ddx5
|
DEAD box helicase 5 |
| chr4_-_133484080 | 0.81 |
ENSMUST00000008024.7
|
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr2_+_130116357 | 0.81 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr9_+_14695933 | 0.79 |
ENSMUST00000034405.11
ENSMUST00000115632.10 ENSMUST00000147305.2 |
Mre11a
|
MRE11A homolog A, double strand break repair nuclease |
| chr3_-_107992662 | 0.78 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr17_-_45883421 | 0.75 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr15_-_38079089 | 0.71 |
ENSMUST00000110336.4
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr9_-_14695801 | 0.70 |
ENSMUST00000214979.2
ENSMUST00000216037.2 ENSMUST00000214456.2 |
Ankrd49
|
ankyrin repeat domain 49 |
| chr2_+_14234198 | 0.67 |
ENSMUST00000028045.4
|
Mrc1
|
mannose receptor, C type 1 |
| chr19_+_9995629 | 0.65 |
ENSMUST00000131407.2
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr11_+_101207021 | 0.64 |
ENSMUST00000142640.8
ENSMUST00000019470.14 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr17_+_47922497 | 0.63 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr1_-_181039509 | 0.62 |
ENSMUST00000162819.9
ENSMUST00000237749.2 |
Wdr26
|
WD repeat domain 26 |
| chr6_+_17463925 | 0.61 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr10_+_36850532 | 0.61 |
ENSMUST00000019911.14
ENSMUST00000105510.2 |
Hdac2
|
histone deacetylase 2 |
| chr17_+_71511642 | 0.61 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr5_+_77099229 | 0.60 |
ENSMUST00000141687.2
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr4_+_21848039 | 0.59 |
ENSMUST00000098238.9
ENSMUST00000108229.2 |
Pnisr
|
PNN interacting serine/arginine-rich |
| chr2_+_18059982 | 0.59 |
ENSMUST00000028076.15
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr19_+_46044972 | 0.59 |
ENSMUST00000111899.8
ENSMUST00000099392.10 ENSMUST00000062322.11 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr4_+_32657105 | 0.59 |
ENSMUST00000071642.11
ENSMUST00000178134.2 |
Mdn1
|
midasin AAA ATPase 1 |
| chr5_+_77122530 | 0.58 |
ENSMUST00000101087.10
ENSMUST00000120550.2 |
Srp72
|
signal recognition particle 72 |
| chr17_+_71923210 | 0.57 |
ENSMUST00000047086.10
|
Wdr43
|
WD repeat domain 43 |
| chr17_+_35219941 | 0.53 |
ENSMUST00000087315.14
|
Vars
|
valyl-tRNA synthetase |
| chr15_-_44651411 | 0.53 |
ENSMUST00000090057.6
ENSMUST00000110269.8 ENSMUST00000228639.2 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr17_+_35219998 | 0.53 |
ENSMUST00000173584.8
|
Vars
|
valyl-tRNA synthetase |
| chr3_+_145281941 | 0.52 |
ENSMUST00000199033.5
ENSMUST00000098534.9 ENSMUST00000200574.5 ENSMUST00000196413.5 ENSMUST00000197604.3 |
Znhit6
|
zinc finger, HIT type 6 |
| chr2_+_83474779 | 0.51 |
ENSMUST00000081591.7
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr7_-_144493560 | 0.51 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr15_+_81695615 | 0.51 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chr16_-_43709968 | 0.51 |
ENSMUST00000023387.14
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr6_+_83142902 | 0.50 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr18_-_60981981 | 0.49 |
ENSMUST00000177172.8
ENSMUST00000175934.8 ENSMUST00000176630.8 |
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chrX_+_35592006 | 0.47 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr12_+_8724681 | 0.45 |
ENSMUST00000168361.8
ENSMUST00000169750.8 ENSMUST00000163730.8 |
Pum2
|
pumilio RNA-binding family member 2 |
| chr4_-_129636073 | 0.45 |
ENSMUST00000066257.6
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr4_-_116263183 | 0.45 |
ENSMUST00000123072.8
ENSMUST00000144281.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr11_+_106107752 | 0.44 |
ENSMUST00000021046.6
|
Ddx42
|
DEAD box helicase 42 |
| chr6_+_125073108 | 0.44 |
ENSMUST00000112390.8
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr1_-_121495623 | 0.43 |
ENSMUST00000001724.12
|
Ddx18
|
DEAD box helicase 18 |
| chr15_-_102533870 | 0.43 |
ENSMUST00000184485.8
ENSMUST00000185070.8 ENSMUST00000184616.8 ENSMUST00000108828.9 |
Atf7
|
activating transcription factor 7 |
| chr5_+_66833434 | 0.41 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr11_-_118181044 | 0.41 |
ENSMUST00000106296.9
ENSMUST00000092382.10 |
Usp36
|
ubiquitin specific peptidase 36 |
| chr14_-_118943591 | 0.41 |
ENSMUST00000036554.14
ENSMUST00000166646.2 |
Abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr12_+_59178072 | 0.40 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr3_+_135531409 | 0.40 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr11_-_76108316 | 0.40 |
ENSMUST00000102500.5
|
Gemin4
|
gem nuclear organelle associated protein 4 |
| chr4_+_109263820 | 0.39 |
ENSMUST00000124209.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr3_+_135531834 | 0.38 |
ENSMUST00000029810.6
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr6_+_17463819 | 0.38 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr16_+_35590745 | 0.37 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr4_+_44756553 | 0.37 |
ENSMUST00000107824.9
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr11_+_106680062 | 0.36 |
ENSMUST00000103068.10
ENSMUST00000018516.11 |
Cep95
|
centrosomal protein 95 |
| chr11_+_100751272 | 0.35 |
ENSMUST00000107357.4
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr14_+_30741082 | 0.34 |
ENSMUST00000112098.11
ENSMUST00000112095.8 ENSMUST00000112106.8 ENSMUST00000146325.8 |
Pbrm1
|
polybromo 1 |
| chrX_-_92675719 | 0.34 |
ENSMUST00000006856.3
|
Pola1
|
polymerase (DNA directed), alpha 1 |
| chr13_-_76091931 | 0.34 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr11_+_70735751 | 0.34 |
ENSMUST00000177731.8
ENSMUST00000108533.10 ENSMUST00000081362.13 ENSMUST00000178245.2 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr13_-_63712349 | 0.34 |
ENSMUST00000192155.6
|
Ptch1
|
patched 1 |
| chr11_-_118180895 | 0.33 |
ENSMUST00000144153.8
|
Usp36
|
ubiquitin specific peptidase 36 |
| chr2_+_115412148 | 0.32 |
ENSMUST00000166472.8
ENSMUST00000110918.3 |
Cdin1
|
CDAN1 interacting nuclease 1 |
| chr11_-_51647204 | 0.32 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr9_+_107464841 | 0.32 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr16_+_18695787 | 0.32 |
ENSMUST00000120532.9
ENSMUST00000004222.14 |
Hira
|
histone cell cycle regulator |
| chr17_-_28705055 | 0.30 |
ENSMUST00000233870.2
|
Fkbp5
|
FK506 binding protein 5 |
| chr4_-_148529187 | 0.28 |
ENSMUST00000051633.3
|
Ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chrX_-_94488394 | 0.28 |
ENSMUST00000084535.6
|
Amer1
|
APC membrane recruitment 1 |
| chr12_+_17594795 | 0.28 |
ENSMUST00000171737.3
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr5_+_35156454 | 0.28 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_-_166237341 | 0.27 |
ENSMUST00000135673.8
ENSMUST00000169324.8 ENSMUST00000128861.3 |
Pogk
|
pogo transposable element with KRAB domain |
| chr7_-_29204812 | 0.27 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr19_-_29790352 | 0.27 |
ENSMUST00000099525.5
|
Ranbp6
|
RAN binding protein 6 |
| chr12_+_59178258 | 0.26 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr9_+_43978290 | 0.26 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr3_+_41519289 | 0.26 |
ENSMUST00000168086.7
|
Jade1
|
jade family PHD finger 1 |
| chr11_-_106107132 | 0.25 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr18_+_32970278 | 0.25 |
ENSMUST00000053663.11
|
Wdr36
|
WD repeat domain 36 |
| chr9_+_40712562 | 0.24 |
ENSMUST00000117557.8
|
Hspa8
|
heat shock protein 8 |
| chr8_+_91555449 | 0.23 |
ENSMUST00000109614.9
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chrX_-_36127891 | 0.23 |
ENSMUST00000115249.10
ENSMUST00000115248.10 |
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr5_+_31855394 | 0.22 |
ENSMUST00000063813.11
ENSMUST00000071531.12 ENSMUST00000131995.7 ENSMUST00000114507.10 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr12_+_71183622 | 0.22 |
ENSMUST00000149564.8
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr18_+_32970363 | 0.21 |
ENSMUST00000166214.9
|
Wdr36
|
WD repeat domain 36 |
| chr3_+_135531548 | 0.21 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr2_+_48839505 | 0.21 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr5_-_108922819 | 0.20 |
ENSMUST00000200159.2
ENSMUST00000212212.2 |
Rnf212
|
ring finger protein 212 |
| chr1_-_52539395 | 0.19 |
ENSMUST00000186764.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr5_+_120254530 | 0.18 |
ENSMUST00000031590.12
|
Rbm19
|
RNA binding motif protein 19 |
| chr4_+_132001680 | 0.18 |
ENSMUST00000030731.11
ENSMUST00000105963.2 |
Taf12
|
TATA-box binding protein associated factor 12 |
| chr14_+_4230658 | 0.18 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr9_+_110162470 | 0.17 |
ENSMUST00000198761.5
ENSMUST00000197630.3 |
Scap
|
SREBF chaperone |
| chr15_+_80556023 | 0.17 |
ENSMUST00000023044.7
|
Fam83f
|
family with sequence similarity 83, member F |
| chr5_+_77099154 | 0.17 |
ENSMUST00000031160.16
ENSMUST00000120912.8 ENSMUST00000117536.8 |
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr19_+_26600820 | 0.17 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_127345909 | 0.16 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr2_-_118380149 | 0.16 |
ENSMUST00000090219.13
|
Bmf
|
BCL2 modifying factor |
| chr17_-_45884179 | 0.16 |
ENSMUST00000165127.8
ENSMUST00000166469.8 ENSMUST00000024739.14 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr11_-_120238917 | 0.15 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr11_+_94520567 | 0.15 |
ENSMUST00000021239.7
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr12_-_32000169 | 0.15 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr5_-_108943211 | 0.15 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr10_+_69048506 | 0.14 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr12_+_69418886 | 0.14 |
ENSMUST00000050063.9
|
Arf6
|
ADP-ribosylation factor 6 |
| chr6_+_49296208 | 0.13 |
ENSMUST00000055559.8
ENSMUST00000114491.2 |
Ccdc126
|
coiled-coil domain containing 126 |
| chr4_-_156077061 | 0.12 |
ENSMUST00000052185.5
|
B3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase, polypeptide 6 |
| chr2_-_168048984 | 0.12 |
ENSMUST00000088001.6
|
Adnp
|
activity-dependent neuroprotective protein |
| chr14_-_20718337 | 0.11 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chr7_-_68398989 | 0.11 |
ENSMUST00000048068.15
|
Arrdc4
|
arrestin domain containing 4 |
| chr4_-_124587340 | 0.10 |
ENSMUST00000030738.8
|
Utp11
|
UTP11 small subunit processome component |
| chr3_-_108117754 | 0.09 |
ENSMUST00000117784.8
ENSMUST00000119650.8 ENSMUST00000117409.8 |
Atxn7l2
|
ataxin 7-like 2 |
| chr9_-_110483210 | 0.09 |
ENSMUST00000196488.5
ENSMUST00000133191.8 ENSMUST00000167320.8 |
Nbeal2
|
neurobeachin-like 2 |
| chr11_-_51891575 | 0.09 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr9_+_110162345 | 0.09 |
ENSMUST00000198976.5
|
Scap
|
SREBF chaperone |
| chr16_-_44978986 | 0.09 |
ENSMUST00000180636.8
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr17_-_47922374 | 0.09 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr11_+_67689094 | 0.08 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr8_-_37081091 | 0.08 |
ENSMUST00000033923.14
|
Dlc1
|
deleted in liver cancer 1 |
| chr19_+_46045675 | 0.08 |
ENSMUST00000126127.8
ENSMUST00000147640.2 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr15_-_51728893 | 0.08 |
ENSMUST00000022925.10
|
Eif3h
|
eukaryotic translation initiation factor 3, subunit H |
| chr17_-_28705082 | 0.08 |
ENSMUST00000079413.11
|
Fkbp5
|
FK506 binding protein 5 |
| chr4_+_44756608 | 0.08 |
ENSMUST00000143385.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr2_+_27405169 | 0.08 |
ENSMUST00000113952.10
|
Wdr5
|
WD repeat domain 5 |
| chr11_+_74721733 | 0.07 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr14_+_30741115 | 0.06 |
ENSMUST00000112094.8
ENSMUST00000144009.2 |
Pbrm1
|
polybromo 1 |
| chr12_-_32000209 | 0.06 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr16_+_43710163 | 0.05 |
ENSMUST00000132859.8
ENSMUST00000178400.9 |
Ccdc191
|
coiled-coil domain containing 191 |
| chr16_-_44978546 | 0.05 |
ENSMUST00000114600.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr1_-_186438177 | 0.04 |
ENSMUST00000045288.14
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr19_-_29782907 | 0.04 |
ENSMUST00000175726.8
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr2_-_48839218 | 0.03 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr12_+_24701273 | 0.03 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr9_+_43978369 | 0.03 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr11_-_100650768 | 0.03 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr16_+_23247883 | 0.02 |
ENSMUST00000038730.7
|
Rtp1
|
receptor transporter protein 1 |
| chr9_-_121324744 | 0.02 |
ENSMUST00000035120.6
|
Cck
|
cholecystokinin |
| chr16_-_44978929 | 0.02 |
ENSMUST00000181177.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr9_+_58041300 | 0.02 |
ENSMUST00000134955.8
ENSMUST00000147134.8 ENSMUST00000170397.8 |
Stra6
|
stimulated by retinoic acid gene 6 |
| chr7_+_19311212 | 0.02 |
ENSMUST00000108453.2
|
Zfp296
|
zinc finger protein 296 |
| chr1_-_186437760 | 0.02 |
ENSMUST00000195201.2
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr16_-_33787399 | 0.00 |
ENSMUST00000023510.7
|
Umps
|
uridine monophosphate synthetase |
| chr2_+_90865958 | 0.00 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr3_+_88439616 | 0.00 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.3 | 2.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 1.9 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.3 | 0.9 | GO:0086097 | actin filament uncapping(GO:0051695) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 0.9 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.3 | 5.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.3 | 1.8 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 1.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 | 0.8 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 1.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 1.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 1.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 0.7 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.2 | 1.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 0.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.4 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 0.9 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 2.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 1.5 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.1 | 0.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 1.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.3 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 1.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.3 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.1 | 0.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.2 | GO:0097212 | protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.8 | GO:0006188 | IMP biosynthetic process(GO:0006188) |
| 0.1 | 0.5 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.2 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.4 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.9 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.2 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.0 | 0.4 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.3 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.4 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.8 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.4 | GO:0006855 | drug transmembrane transport(GO:0006855) prostaglandin secretion(GO:0032310) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:1905006 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.7 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 1.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.7 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 1.5 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.3 | 2.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 1.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 0.9 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 0.9 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.2 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 2.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 2.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.6 | 1.7 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.4 | 5.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 1.9 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.3 | 2.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 0.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 2.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 0.6 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 0.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 1.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.9 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.6 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.9 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 5.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |