Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Crx_Gsc
Z-value: 0.45
Transcription factors associated with Crx_Gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
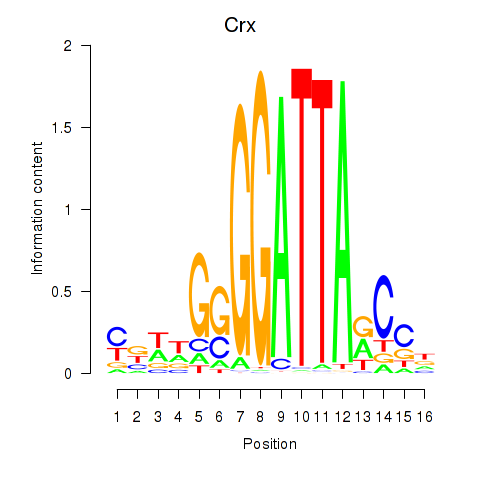
Crx
|
ENSMUSG00000041578.16 | Crx |
|
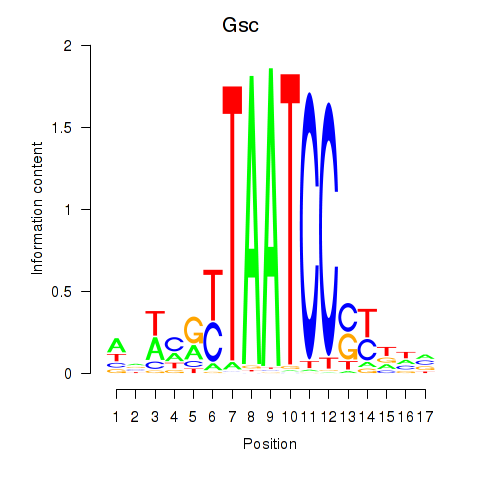
Gsc
|
ENSMUSG00000021095.6 | Gsc |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Crx | mm39_v1_chr7_-_15613769_15613893 | 0.23 | 1.8e-01 | Click! |
| Gsc | mm39_v1_chr12_-_104439589_104439633 | -0.19 | 2.6e-01 | Click! |
Activity profile of Crx_Gsc motif
Sorted Z-values of Crx_Gsc motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Crx_Gsc
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_4755971 | 2.32 |
ENSMUST00000183971.8
ENSMUST00000182173.2 ENSMUST00000182738.8 ENSMUST00000182111.8 ENSMUST00000184143.8 ENSMUST00000182048.2 ENSMUST00000063324.14 |
Cox6b2
|
cytochrome c oxidase subunit 6B2 |
| chr7_-_103492361 | 1.56 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr14_-_87378641 | 1.53 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chr16_+_36004452 | 1.30 |
ENSMUST00000114858.2
|
Cstdc4
|
cystatin domain containing 4 |
| chr4_-_140805613 | 1.26 |
ENSMUST00000030760.15
|
Necap2
|
NECAP endocytosis associated 2 |
| chr13_-_4200627 | 1.21 |
ENSMUST00000110704.9
ENSMUST00000021635.9 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chr11_-_16958647 | 1.06 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr8_+_117822593 | 0.93 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr4_-_63090355 | 0.92 |
ENSMUST00000156618.9
ENSMUST00000030042.3 |
Kif12
|
kinesin family member 12 |
| chr6_+_113508636 | 0.85 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr7_+_99184645 | 0.80 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr14_-_20027219 | 0.77 |
ENSMUST00000055100.14
ENSMUST00000162425.8 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr10_-_93727003 | 0.76 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr11_+_94901104 | 0.67 |
ENSMUST00000124735.2
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr14_-_20319242 | 0.66 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr16_+_33614378 | 0.64 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr10_-_62215631 | 0.61 |
ENSMUST00000143236.8
ENSMUST00000133429.8 ENSMUST00000132926.8 ENSMUST00000116238.9 |
Hk1
|
hexokinase 1 |
| chr11_+_120489358 | 0.58 |
ENSMUST00000093140.5
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr19_-_6178171 | 0.56 |
ENSMUST00000154601.8
ENSMUST00000138931.3 |
Snx15
|
sorting nexin 15 |
| chr7_+_99243812 | 0.54 |
ENSMUST00000162290.2
|
Arrb1
|
arrestin, beta 1 |
| chr10_-_62163444 | 0.52 |
ENSMUST00000139228.8
|
Hk1
|
hexokinase 1 |
| chr1_+_172328768 | 0.52 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr16_-_16687119 | 0.51 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr7_-_44635813 | 0.49 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr6_+_123206802 | 0.49 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr14_+_26722319 | 0.49 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr7_-_44741622 | 0.49 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr19_-_5416339 | 0.49 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr18_-_74340885 | 0.48 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr7_+_65710086 | 0.47 |
ENSMUST00000153609.8
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr4_-_108263873 | 0.45 |
ENSMUST00000184609.2
|
Gpx7
|
glutathione peroxidase 7 |
| chr17_-_26160872 | 0.45 |
ENSMUST00000139226.2
ENSMUST00000097368.10 ENSMUST00000140304.2 ENSMUST00000026823.16 |
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr17_+_36132567 | 0.44 |
ENSMUST00000003635.7
|
Ier3
|
immediate early response 3 |
| chr18_-_73948501 | 0.43 |
ENSMUST00000025439.5
|
Me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr17_-_26160826 | 0.42 |
ENSMUST00000208071.2
|
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr10_+_94034817 | 0.42 |
ENSMUST00000020209.16
ENSMUST00000179990.8 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr6_+_122898764 | 0.42 |
ENSMUST00000060484.9
|
Clec4a1
|
C-type lectin domain family 4, member a1 |
| chrX_-_101232978 | 0.41 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr4_+_130640436 | 0.41 |
ENSMUST00000151698.8
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr11_+_70396070 | 0.41 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr15_-_76524097 | 0.41 |
ENSMUST00000168185.8
|
Tonsl
|
tonsoku-like, DNA repair protein |
| chr11_-_40624200 | 0.41 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr13_+_21995906 | 0.41 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17 |
| chr4_-_116851571 | 0.40 |
ENSMUST00000030446.15
|
Urod
|
uroporphyrinogen decarboxylase |
| chr12_-_101943134 | 0.40 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr5_-_74229021 | 0.40 |
ENSMUST00000119154.8
ENSMUST00000068058.14 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr15_+_72962241 | 0.40 |
ENSMUST00000089765.9
ENSMUST00000134353.2 |
Chrac1
|
chromatin accessibility complex 1 |
| chr5_-_142594549 | 0.40 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr18_-_74340842 | 0.38 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr2_+_120307390 | 0.38 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr11_+_109540201 | 0.38 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr12_-_114621406 | 0.37 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr7_-_44741609 | 0.37 |
ENSMUST00000210734.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr10_+_75778885 | 0.37 |
ENSMUST00000121151.2
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chr14_-_20027112 | 0.37 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr14_-_20027279 | 0.36 |
ENSMUST00000160013.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr7_-_132388643 | 0.36 |
ENSMUST00000065371.14
ENSMUST00000106166.8 |
Fam53b
|
family with sequence similarity 53, member B |
| chr10_+_90412827 | 0.36 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr16_+_33614715 | 0.36 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr11_-_82799186 | 0.36 |
ENSMUST00000103213.10
|
Nle1
|
notchless homolog 1 |
| chr11_-_75330302 | 0.35 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr5_+_114268425 | 0.35 |
ENSMUST00000031587.13
|
Ung
|
uracil DNA glycosylase |
| chr5_-_92426014 | 0.35 |
ENSMUST00000159345.8
ENSMUST00000113102.10 |
Naaa
|
N-acylethanolamine acid amidase |
| chr5_-_5744559 | 0.34 |
ENSMUST00000115426.9
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr5_+_143636977 | 0.34 |
ENSMUST00000110727.2
|
Cyth3
|
cytohesin 3 |
| chr19_-_5416626 | 0.34 |
ENSMUST00000237167.2
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr7_+_92524460 | 0.34 |
ENSMUST00000076052.8
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr8_-_108129829 | 0.34 |
ENSMUST00000003947.9
|
Nqo1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr18_-_60981981 | 0.33 |
ENSMUST00000177172.8
ENSMUST00000175934.8 ENSMUST00000176630.8 |
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr9_-_50528727 | 0.33 |
ENSMUST00000131351.8
ENSMUST00000171462.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr19_+_53128901 | 0.32 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chrX_-_99670174 | 0.31 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chr11_-_109364046 | 0.31 |
ENSMUST00000070872.13
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr11_+_68906737 | 0.31 |
ENSMUST00000021278.14
ENSMUST00000161455.8 ENSMUST00000116359.3 |
Ctc1
|
CTS telomere maintenance complex component 1 |
| chr1_-_152500963 | 0.30 |
ENSMUST00000027760.14
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr7_-_110581652 | 0.29 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr3_-_16060545 | 0.29 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr19_+_53128861 | 0.29 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr1_-_149798090 | 0.29 |
ENSMUST00000111926.9
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr9_+_26941453 | 0.29 |
ENSMUST00000073127.14
ENSMUST00000086198.5 |
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr4_-_140501507 | 0.29 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr15_+_80017315 | 0.29 |
ENSMUST00000023050.9
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr10_+_39009951 | 0.28 |
ENSMUST00000019991.8
|
Tube1
|
tubulin, epsilon 1 |
| chr10_-_86843878 | 0.28 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr10_+_59839142 | 0.28 |
ENSMUST00000164083.4
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr7_+_92524495 | 0.28 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr14_+_51193449 | 0.27 |
ENSMUST00000095925.5
|
Pnp2
|
purine-nucleoside phosphorylase 2 |
| chr11_+_29668563 | 0.27 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr7_-_113875342 | 0.27 |
ENSMUST00000033008.10
|
Psma1
|
proteasome subunit alpha 1 |
| chr6_-_69553484 | 0.26 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr9_+_108820846 | 0.26 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr1_-_189420270 | 0.26 |
ENSMUST00000171929.8
ENSMUST00000165962.2 |
Cenpf
|
centromere protein F |
| chr11_-_69470139 | 0.26 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr18_-_32044877 | 0.26 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr8_-_71326027 | 0.26 |
ENSMUST00000212680.2
|
Ccdc124
|
coiled-coil domain containing 124 |
| chr3_+_138058139 | 0.26 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr8_-_94825556 | 0.26 |
ENSMUST00000034206.6
|
Bbs2
|
Bardet-Biedl syndrome 2 (human) |
| chr8_+_71126012 | 0.25 |
ENSMUST00000146972.3
ENSMUST00000210071.2 ENSMUST00000210987.2 |
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_+_29251602 | 0.25 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr7_-_113875261 | 0.25 |
ENSMUST00000135570.8
|
Psma1
|
proteasome subunit alpha 1 |
| chr7_-_44635740 | 0.25 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr7_+_24584197 | 0.25 |
ENSMUST00000156372.8
ENSMUST00000124035.2 |
Rps19
|
ribosomal protein S19 |
| chr3_+_103767581 | 0.25 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr11_-_116164928 | 0.25 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr10_-_93725619 | 0.25 |
ENSMUST00000181091.8
ENSMUST00000181217.8 ENSMUST00000047910.15 ENSMUST00000180688.2 |
Metap2
|
methionine aminopeptidase 2 |
| chr17_+_26342474 | 0.24 |
ENSMUST00000025014.10
ENSMUST00000236166.2 ENSMUST00000127647.3 |
Mrpl28
|
mitochondrial ribosomal protein L28 |
| chr11_+_101137786 | 0.24 |
ENSMUST00000107282.4
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr6_-_69204417 | 0.24 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr2_+_24275321 | 0.24 |
ENSMUST00000056641.15
ENSMUST00000142522.8 ENSMUST00000131930.2 |
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr5_-_137530214 | 0.24 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr17_-_35351026 | 0.23 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr6_-_69220672 | 0.23 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr11_+_120489247 | 0.23 |
ENSMUST00000026128.10
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr11_-_102907991 | 0.23 |
ENSMUST00000021313.9
|
Dcakd
|
dephospho-CoA kinase domain containing |
| chr3_-_16060491 | 0.23 |
ENSMUST00000108347.3
|
Gm5150
|
predicted gene 5150 |
| chr17_-_13211374 | 0.22 |
ENSMUST00000159551.8
ENSMUST00000160781.8 |
Wtap
|
Wilms tumour 1-associating protein |
| chr10_-_59838878 | 0.22 |
ENSMUST00000182912.2
ENSMUST00000020307.11 ENSMUST00000182898.8 |
Anapc16
|
anaphase promoting complex subunit 16 |
| chr13_+_12410240 | 0.22 |
ENSMUST00000059270.10
|
Heatr1
|
HEAT repeat containing 1 |
| chr8_+_71125876 | 0.22 |
ENSMUST00000034311.15
|
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_-_68713748 | 0.22 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr6_-_113717689 | 0.21 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr17_+_34124078 | 0.21 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr5_-_100307130 | 0.21 |
ENSMUST00000139520.3
|
Tmem150c
|
transmembrane protein 150C |
| chr17_-_28660688 | 0.21 |
ENSMUST00000233291.2
ENSMUST00000153744.2 |
Fkbp5
|
FK506 binding protein 5 |
| chr6_+_117877272 | 0.21 |
ENSMUST00000177743.8
ENSMUST00000167182.8 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr1_+_72346572 | 0.21 |
ENSMUST00000027379.10
|
Xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr14_+_53607470 | 0.21 |
ENSMUST00000103652.5
|
Trav14n-3
|
T cell receptor alpha variable 14N-3 |
| chr5_+_117258185 | 0.20 |
ENSMUST00000111978.8
|
Taok3
|
TAO kinase 3 |
| chr11_+_60030891 | 0.20 |
ENSMUST00000171108.8
ENSMUST00000090806.5 |
Rai1
|
retinoic acid induced 1 |
| chr11_-_109363406 | 0.20 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr11_-_120489172 | 0.20 |
ENSMUST00000026125.3
|
Alyref
|
Aly/REF export factor |
| chr6_+_123206880 | 0.19 |
ENSMUST00000205129.2
|
Clec4n
|
C-type lectin domain family 4, member n |
| chr11_-_116165024 | 0.19 |
ENSMUST00000021133.16
|
Srp68
|
signal recognition particle 68 |
| chr19_+_8861096 | 0.19 |
ENSMUST00000187504.7
|
Lbhd1
|
LBH domain containing 1 |
| chr17_+_34092811 | 0.19 |
ENSMUST00000226885.2
|
Smim40-ps
|
small integral membrane protein 40, pseudogene |
| chr2_-_144112444 | 0.19 |
ENSMUST00000028909.5
|
Snx5
|
sorting nexin 5 |
| chr16_+_38383154 | 0.18 |
ENSMUST00000171687.8
ENSMUST00000002924.15 |
Tmem39a
|
transmembrane protein 39a |
| chr7_-_29892523 | 0.18 |
ENSMUST00000108196.8
|
Capns1
|
calpain, small subunit 1 |
| chr2_-_87838612 | 0.18 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr7_-_110673269 | 0.18 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr5_+_122348140 | 0.18 |
ENSMUST00000196187.5
ENSMUST00000100747.3 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr8_+_4303067 | 0.18 |
ENSMUST00000011981.5
ENSMUST00000208316.2 |
Snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr11_+_90078485 | 0.17 |
ENSMUST00000124877.3
ENSMUST00000153734.2 ENSMUST00000134929.8 |
Gm45716
|
predicted gene 45716 |
| chr13_+_94925382 | 0.17 |
ENSMUST00000046644.7
|
Tbca
|
tubulin cofactor A |
| chr5_+_64316771 | 0.17 |
ENSMUST00000043893.13
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr15_+_82136598 | 0.17 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr18_+_42644552 | 0.17 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr16_-_30900181 | 0.17 |
ENSMUST00000055389.9
|
Xxylt1
|
xyloside xylosyltransferase 1 |
| chr1_+_172302925 | 0.17 |
ENSMUST00000027830.5
|
Slamf9
|
SLAM family member 9 |
| chr11_+_101137231 | 0.16 |
ENSMUST00000122006.8
ENSMUST00000151830.2 |
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr5_-_145103841 | 0.16 |
ENSMUST00000031628.10
|
Ptcd1
|
pentatricopeptide repeat domain 1 |
| chr15_-_78058214 | 0.16 |
ENSMUST00000016781.8
|
Ift27
|
intraflagellar transport 27 |
| chr8_+_79236051 | 0.16 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr13_-_74918701 | 0.16 |
ENSMUST00000223126.2
|
Cast
|
calpastatin |
| chr12_+_85645801 | 0.16 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr10_-_79875479 | 0.16 |
ENSMUST00000004786.10
|
Polr2e
|
polymerase (RNA) II (DNA directed) polypeptide E |
| chr12_+_101942222 | 0.16 |
ENSMUST00000047357.10
|
Cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr15_+_76832681 | 0.15 |
ENSMUST00000071792.7
ENSMUST00000230274.2 |
1110038F14Rik
|
RIKEN cDNA 1110038F14 gene |
| chr12_-_115157739 | 0.15 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr2_+_67276338 | 0.15 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr5_-_137529465 | 0.15 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr7_-_43182504 | 0.15 |
ENSMUST00000004728.12
|
Cd33
|
CD33 antigen |
| chr10_+_80097290 | 0.14 |
ENSMUST00000156935.8
|
Dazap1
|
DAZ associated protein 1 |
| chr9_+_73020708 | 0.14 |
ENSMUST00000169399.8
ENSMUST00000034738.14 |
Rsl24d1
|
ribosomal L24 domain containing 1 |
| chr13_-_74918745 | 0.14 |
ENSMUST00000223033.2
ENSMUST00000222588.2 |
Cast
|
calpastatin |
| chr3_+_105778174 | 0.14 |
ENSMUST00000164730.2
ENSMUST00000010279.10 |
Adora3
Tmigd3
|
adenosine A3 receptor transmembrane and immunoglobulin domain containing 3 |
| chr5_+_136023649 | 0.14 |
ENSMUST00000111142.9
ENSMUST00000111145.10 ENSMUST00000111144.8 ENSMUST00000199239.5 ENSMUST00000005072.10 ENSMUST00000130345.2 |
Dtx2
|
deltex 2, E3 ubiquitin ligase |
| chrX_-_149371067 | 0.14 |
ENSMUST00000140207.8
ENSMUST00000112719.2 ENSMUST00000112727.10 ENSMUST00000112721.10 ENSMUST00000026303.16 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr4_+_88673105 | 0.14 |
ENSMUST00000094992.2
|
Gm13271
|
predicted gene 13271 |
| chr3_+_92223927 | 0.14 |
ENSMUST00000061038.4
|
Sprr2b
|
small proline-rich protein 2B |
| chr1_+_74193138 | 0.13 |
ENSMUST00000027372.8
ENSMUST00000106899.4 |
Cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr11_+_83553400 | 0.13 |
ENSMUST00000019074.4
|
Ccl4
|
chemokine (C-C motif) ligand 4 |
| chr15_-_64254754 | 0.13 |
ENSMUST00000177374.8
ENSMUST00000110114.10 ENSMUST00000110115.9 ENSMUST00000023008.16 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr12_-_114901026 | 0.13 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr11_+_119205613 | 0.13 |
ENSMUST00000053245.7
|
Card14
|
caspase recruitment domain family, member 14 |
| chr8_-_62576140 | 0.13 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr10_+_77458109 | 0.13 |
ENSMUST00000174510.8
ENSMUST00000172813.2 |
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr11_+_3913970 | 0.13 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr7_+_110628158 | 0.13 |
ENSMUST00000005749.6
|
Ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr7_-_103778992 | 0.12 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr2_+_152468850 | 0.12 |
ENSMUST00000000369.4
ENSMUST00000150913.2 |
Rem1
|
rad and gem related GTP binding protein 1 |
| chr17_+_35960600 | 0.12 |
ENSMUST00000171166.3
|
Sfta2
|
surfactant associated 2 |
| chr14_-_20596508 | 0.12 |
ENSMUST00000161989.2
|
Ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isoform |
| chr15_+_76832920 | 0.12 |
ENSMUST00000229229.2
|
1110038F14Rik
|
RIKEN cDNA 1110038F14 gene |
| chr1_-_181985663 | 0.12 |
ENSMUST00000169123.4
|
Vmn1r1
|
vomeronasal 1 receptor 1 |
| chr19_-_7688628 | 0.12 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr4_-_149822480 | 0.12 |
ENSMUST00000105687.9
ENSMUST00000054459.11 ENSMUST00000103208.2 |
Tmem201
|
transmembrane protein 201 |
| chr7_-_43182595 | 0.12 |
ENSMUST00000205503.2
|
Cd33
|
CD33 antigen |
| chr9_-_62895197 | 0.11 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1 |
| chr1_-_173018204 | 0.11 |
ENSMUST00000215878.2
ENSMUST00000201132.3 |
Olfr1406
|
olfactory receptor 1406 |
| chr10_-_39009910 | 0.11 |
ENSMUST00000135785.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr7_-_110581376 | 0.11 |
ENSMUST00000154466.2
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr19_+_5074070 | 0.11 |
ENSMUST00000025826.7
ENSMUST00000237371.2 ENSMUST00000235416.2 |
Slc29a2
|
solute carrier family 29 (nucleoside transporters), member 2 |
| chr5_-_65248927 | 0.11 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr13_-_64645606 | 0.11 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr5_-_123859153 | 0.11 |
ENSMUST00000196282.5
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr7_-_4535403 | 0.11 |
ENSMUST00000094897.5
|
Dnaaf3
|
dynein, axonemal assembly factor 3 |
| chr11_+_95925711 | 0.11 |
ENSMUST00000006217.10
ENSMUST00000107700.4 |
Snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr1_-_192453521 | 0.11 |
ENSMUST00000128619.8
ENSMUST00000044190.12 |
Hhat
|
hedgehog acyltransferase |
| chr7_-_119078472 | 0.11 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0009753 | sesquiterpenoid metabolic process(GO:0006714) response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 1.1 | GO:0060305 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.3 | 0.8 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.2 | 1.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 0.6 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.7 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.1 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.4 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.1 | 0.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.2 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.4 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 0.2 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 | 0.8 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 1.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.3 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.3 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.2 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.4 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.2 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.4 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.4 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0032202 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.4 | GO:0048149 | adult feeding behavior(GO:0008343) behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.3 | GO:0007343 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.5 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0003142 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) protein lipoylation(GO:0009249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 2.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.4 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 1.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 1.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0031597 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 1.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 1.6 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.2 | 0.8 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.4 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.3 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.4 | GO:0030942 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.0 | 0.1 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.4 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 0.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |