Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Cxxc1
Z-value: 0.32
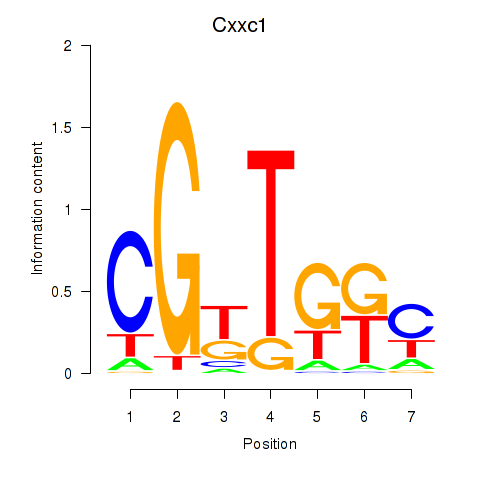
Transcription factors associated with Cxxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cxxc1
|
ENSMUSG00000024560.8 | Cxxc1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cxxc1 | mm39_v1_chr18_+_74349189_74349217 | -0.20 | 2.3e-01 | Click! |
Activity profile of Cxxc1 motif
Sorted Z-values of Cxxc1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Cxxc1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_96175970 | 0.31 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chr15_-_38518406 | 0.28 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr11_+_70410445 | 0.25 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr19_+_38919477 | 0.23 |
ENSMUST00000145051.2
|
Hells
|
helicase, lymphoid specific |
| chr15_-_38518458 | 0.21 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr7_+_55700089 | 0.21 |
ENSMUST00000164095.3
|
Herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr19_-_19088543 | 0.21 |
ENSMUST00000112832.8
|
Rorb
|
RAR-related orphan receptor beta |
| chr10_+_75790348 | 0.20 |
ENSMUST00000099577.4
|
Gm5134
|
predicted gene 5134 |
| chr10_+_80085275 | 0.19 |
ENSMUST00000020361.7
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr18_-_43610829 | 0.18 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr11_-_17903861 | 0.18 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr8_+_93084253 | 0.16 |
ENSMUST00000210246.2
ENSMUST00000034184.12 |
Irx5
|
Iroquois homeobox 5 |
| chr1_+_86354045 | 0.14 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr10_+_127894843 | 0.14 |
ENSMUST00000084771.3
|
Ptges3
|
prostaglandin E synthase 3 |
| chr9_-_78016302 | 0.14 |
ENSMUST00000001402.14
|
Fbxo9
|
f-box protein 9 |
| chr10_+_127894816 | 0.13 |
ENSMUST00000052798.14
|
Ptges3
|
prostaglandin E synthase 3 |
| chr5_+_14075281 | 0.13 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr11_+_60428788 | 0.12 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr11_-_88609048 | 0.12 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_+_45546365 | 0.11 |
ENSMUST00000069772.16
ENSMUST00000210503.2 ENSMUST00000209350.2 |
Tmem143
|
transmembrane protein 143 |
| chr4_-_42853888 | 0.11 |
ENSMUST00000107979.2
|
Fam205a1
|
family with sequence similarity 205, member A1 |
| chr3_-_41696906 | 0.11 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr8_+_84441806 | 0.11 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a |
| chr1_+_59296065 | 0.10 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr14_-_62718581 | 0.10 |
ENSMUST00000186010.8
|
Gm4131
|
predicted gene 4131 |
| chr5_+_65548840 | 0.10 |
ENSMUST00000122026.6
ENSMUST00000031101.10 ENSMUST00000200374.2 |
Lias
|
lipoic acid synthetase |
| chr13_-_55979191 | 0.10 |
ENSMUST00000021968.7
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr8_+_84442133 | 0.10 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a |
| chr11_-_88608958 | 0.09 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr5_+_25721059 | 0.09 |
ENSMUST00000045016.9
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr8_+_84441854 | 0.09 |
ENSMUST00000172396.8
|
Ddx39a
|
DEAD box helicase 39a |
| chr3_-_146205429 | 0.08 |
ENSMUST00000029839.11
|
Spata1
|
spermatogenesis associated 1 |
| chr1_-_14989059 | 0.08 |
ENSMUST00000235071.2
ENSMUST00000041447.5 |
Trpa1
|
transient receptor potential cation channel, subfamily A, member 1 |
| chr6_-_135287372 | 0.08 |
ENSMUST00000050471.4
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr9_+_64940004 | 0.08 |
ENSMUST00000167773.2
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr18_+_37138256 | 0.08 |
ENSMUST00000115658.6
|
Pcdha11
|
protocadherin alpha 11 |
| chr9_-_78015866 | 0.07 |
ENSMUST00000162625.2
ENSMUST00000159099.8 ENSMUST00000085311.13 |
Fbxo9
|
f-box protein 9 |
| chr11_-_85125889 | 0.07 |
ENSMUST00000018625.10
|
Appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr7_-_49286594 | 0.07 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr1_-_5987617 | 0.07 |
ENSMUST00000044180.5
|
Npbwr1
|
neuropeptides B/W receptor 1 |
| chr2_+_112092271 | 0.06 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr10_-_51507527 | 0.04 |
ENSMUST00000219286.2
ENSMUST00000020062.4 ENSMUST00000218684.2 |
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr4_+_108857967 | 0.03 |
ENSMUST00000106644.9
|
Nrd1
|
nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
| chr9_-_107483855 | 0.03 |
ENSMUST00000073448.12
ENSMUST00000194606.2 ENSMUST00000195662.6 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr2_+_151753119 | 0.03 |
ENSMUST00000028955.6
|
Angpt4
|
angiopoietin 4 |
| chr2_+_146854916 | 0.02 |
ENSMUST00000028921.6
|
Xrn2
|
5'-3' exoribonuclease 2 |
| chr19_-_36896999 | 0.02 |
ENSMUST00000238948.2
ENSMUST00000057337.9 |
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chrX_+_5959507 | 0.01 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr17_+_28076209 | 0.01 |
ENSMUST00000130810.2
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr5_-_110957933 | 0.01 |
ENSMUST00000031490.11
|
Ulk1
|
unc-51 like kinase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |