Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Dbp
Z-value: 0.83
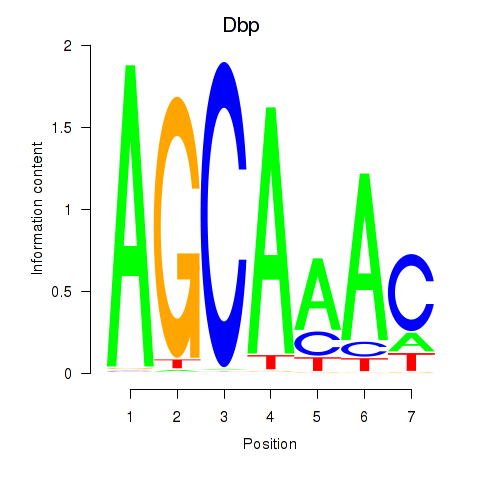
Transcription factors associated with Dbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dbp
|
ENSMUSG00000059824.13 | Dbp |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dbp | mm39_v1_chr7_+_45354512_45354735 | -0.06 | 7.3e-01 | Click! |
Activity profile of Dbp motif
Sorted Z-values of Dbp motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Dbp
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_115604321 | 2.06 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr5_-_77262968 | 1.77 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr17_-_31363245 | 1.72 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr7_-_126651847 | 1.64 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr6_-_141801897 | 1.57 |
ENSMUST00000165990.8
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr12_-_84497718 | 1.52 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr3_-_113166153 | 1.41 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr6_-_141801918 | 1.38 |
ENSMUST00000163678.2
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr1_-_150341911 | 1.10 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr3_+_59989282 | 1.05 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr8_+_105858432 | 0.95 |
ENSMUST00000161289.2
|
Ces4a
|
carboxylesterase 4A |
| chr2_+_67948057 | 0.83 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr14_+_120513106 | 0.82 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr12_+_8027767 | 0.82 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr12_+_8027640 | 0.81 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr15_+_41694317 | 0.81 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr19_+_26725589 | 0.81 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_-_93889483 | 0.80 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr11_-_11848044 | 0.76 |
ENSMUST00000066237.10
|
Ddc
|
dopa decarboxylase |
| chr2_-_77533596 | 0.76 |
ENSMUST00000171063.8
|
Zfp385b
|
zinc finger protein 385B |
| chr11_-_84058292 | 0.76 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr10_+_87696339 | 0.73 |
ENSMUST00000121161.8
|
Igf1
|
insulin-like growth factor 1 |
| chr11_-_11848107 | 0.71 |
ENSMUST00000178704.8
|
Ddc
|
dopa decarboxylase |
| chr8_-_85500998 | 0.70 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr17_-_47040083 | 0.69 |
ENSMUST00000002846.9
|
Gnmt
|
glycine N-methyltransferase |
| chr5_-_51711237 | 0.67 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr1_+_133292898 | 0.66 |
ENSMUST00000129213.2
|
Etnk2
|
ethanolamine kinase 2 |
| chr14_+_120513076 | 0.65 |
ENSMUST00000088419.13
|
Mbnl2
|
muscleblind like splicing factor 2 |
| chr8_-_85500010 | 0.65 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr18_-_39620115 | 0.64 |
ENSMUST00000097592.9
ENSMUST00000115571.8 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr3_+_57643477 | 0.63 |
ENSMUST00000041826.14
ENSMUST00000198510.5 ENSMUST00000200497.5 ENSMUST00000198214.5 ENSMUST00000200600.5 ENSMUST00000198249.5 ENSMUST00000199041.2 |
Rnf13
|
ring finger protein 13 |
| chr8_+_46080840 | 0.62 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_-_11840394 | 0.61 |
ENSMUST00000109659.9
|
Ddc
|
dopa decarboxylase |
| chr3_-_89230190 | 0.61 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr10_+_87695886 | 0.60 |
ENSMUST00000062862.13
|
Igf1
|
insulin-like growth factor 1 |
| chr1_-_66856695 | 0.59 |
ENSMUST00000068168.10
ENSMUST00000113987.2 |
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr14_-_61677258 | 0.59 |
ENSMUST00000022496.9
|
Kpna3
|
karyopherin (importin) alpha 3 |
| chr2_-_72810782 | 0.58 |
ENSMUST00000102689.10
|
Sp3
|
trans-acting transcription factor 3 |
| chr3_+_137983250 | 0.56 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr4_-_109013807 | 0.55 |
ENSMUST00000161363.2
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr2_+_164245114 | 0.51 |
ENSMUST00000017151.2
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr2_+_107120934 | 0.51 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr9_+_18203421 | 0.50 |
ENSMUST00000001825.9
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chr2_+_145627900 | 0.50 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr8_+_46080746 | 0.50 |
ENSMUST00000145458.9
ENSMUST00000134321.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr9_+_51958453 | 0.49 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr14_+_79753055 | 0.49 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr12_+_65012564 | 0.49 |
ENSMUST00000066296.9
ENSMUST00000223166.2 |
Togaram1
|
TOG array regulator of axonemal microtubules 1 |
| chr6_+_134807097 | 0.47 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr10_+_98943999 | 0.46 |
ENSMUST00000161240.4
|
Galnt4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr2_+_68966125 | 0.45 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr1_+_177272215 | 0.45 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr5_-_51711204 | 0.45 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr19_+_26582450 | 0.45 |
ENSMUST00000176769.9
ENSMUST00000208163.2 ENSMUST00000025862.15 ENSMUST00000176030.8 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_66238313 | 0.44 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr13_+_41013230 | 0.43 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr3_-_132940695 | 0.43 |
ENSMUST00000161932.2
|
Arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr1_-_165762469 | 0.43 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr3_+_146276147 | 0.43 |
ENSMUST00000199489.5
|
Uox
|
urate oxidase |
| chr6_-_35110560 | 0.42 |
ENSMUST00000202143.4
ENSMUST00000114993.9 ENSMUST00000114989.9 ENSMUST00000044163.10 ENSMUST00000202417.2 |
Cnot4
|
CCR4-NOT transcription complex, subunit 4 |
| chr12_+_76353835 | 0.41 |
ENSMUST00000220321.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chrX_-_71699740 | 0.40 |
ENSMUST00000055966.13
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr19_-_34856853 | 0.40 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chrX_+_21581135 | 0.40 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr15_-_82796308 | 0.40 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr17_+_44114894 | 0.39 |
ENSMUST00000044895.13
|
Rcan2
|
regulator of calcineurin 2 |
| chr7_+_45434755 | 0.39 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr3_-_132940647 | 0.39 |
ENSMUST00000147041.10
ENSMUST00000161022.9 |
Arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr5_-_36771074 | 0.39 |
ENSMUST00000132383.6
ENSMUST00000174019.2 |
D5Ertd579e
Gm42936
|
DNA segment, Chr 5, ERATO Doi 579, expressed predicted gene 42936 |
| chr15_-_44291226 | 0.39 |
ENSMUST00000227843.2
|
Nudcd1
|
NudC domain containing 1 |
| chr10_+_67021509 | 0.38 |
ENSMUST00000173689.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr9_+_14411921 | 0.38 |
ENSMUST00000213913.2
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr4_+_97665843 | 0.38 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr12_+_84498196 | 0.38 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr2_-_65068960 | 0.38 |
ENSMUST00000112429.9
ENSMUST00000102726.8 ENSMUST00000112430.8 |
Cobll1
|
Cobl-like 1 |
| chr15_-_44291699 | 0.38 |
ENSMUST00000038719.8
|
Nudcd1
|
NudC domain containing 1 |
| chr8_+_47192767 | 0.37 |
ENSMUST00000034041.9
ENSMUST00000208507.2 ENSMUST00000207105.2 |
Irf2
|
interferon regulatory factor 2 |
| chr9_+_14411907 | 0.37 |
ENSMUST00000004200.9
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr4_-_57916283 | 0.37 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chrX_+_36059274 | 0.37 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr15_-_39720855 | 0.36 |
ENSMUST00000022915.11
ENSMUST00000110306.9 |
Dpys
|
dihydropyrimidinase |
| chr4_+_43669266 | 0.36 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr1_+_24717968 | 0.35 |
ENSMUST00000095062.10
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr2_+_29968225 | 0.35 |
ENSMUST00000150770.8
|
Pkn3
|
protein kinase N3 |
| chr3_-_148696155 | 0.35 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr10_+_128626772 | 0.34 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr7_-_19595221 | 0.34 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr17_-_12894716 | 0.34 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr11_-_96714813 | 0.33 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr5_+_102992873 | 0.33 |
ENSMUST00000070000.6
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr3_-_57483330 | 0.33 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr2_+_153716958 | 0.33 |
ENSMUST00000028983.3
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr1_+_33947250 | 0.33 |
ENSMUST00000183034.5
|
Dst
|
dystonin |
| chr3_-_51184895 | 0.32 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr11_-_96714950 | 0.32 |
ENSMUST00000169828.8
ENSMUST00000126949.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr5_-_92190489 | 0.31 |
ENSMUST00000113140.5
ENSMUST00000113143.8 |
Cdkl2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr4_+_97665992 | 0.31 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr13_+_113931038 | 0.31 |
ENSMUST00000091201.7
|
Arl15
|
ADP-ribosylation factor-like 15 |
| chr2_-_65068917 | 0.30 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr5_+_42225303 | 0.30 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr8_-_41469786 | 0.30 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr17_-_45903188 | 0.29 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr5_+_98328723 | 0.29 |
ENSMUST00000112959.4
|
Prdm8
|
PR domain containing 8 |
| chrX_-_43879022 | 0.29 |
ENSMUST00000115056.2
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr10_+_62935430 | 0.28 |
ENSMUST00000044059.5
|
Atoh7
|
atonal bHLH transcription factor 7 |
| chr3_+_60436163 | 0.28 |
ENSMUST00000194201.6
ENSMUST00000193517.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr8_-_23143422 | 0.28 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
| chr12_+_86994117 | 0.28 |
ENSMUST00000187814.7
ENSMUST00000038369.11 |
Cipc
|
CLOCK interacting protein, circadian |
| chr7_+_131150484 | 0.28 |
ENSMUST00000183219.8
|
Hmx2
|
H6 homeobox 2 |
| chr8_-_79539838 | 0.28 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_105505772 | 0.27 |
ENSMUST00000111085.8
|
Pax6
|
paired box 6 |
| chr11_-_77678573 | 0.27 |
ENSMUST00000092883.3
|
Gm10277
|
predicted gene 10277 |
| chr7_-_16761732 | 0.27 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr1_-_43866910 | 0.27 |
ENSMUST00000153317.6
ENSMUST00000128261.2 ENSMUST00000126008.8 ENSMUST00000139451.8 |
Uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr6_+_134807170 | 0.27 |
ENSMUST00000111937.2
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr10_-_89369432 | 0.27 |
ENSMUST00000105297.2
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr12_-_65012270 | 0.26 |
ENSMUST00000222508.2
|
Klhl28
|
kelch-like 28 |
| chr1_+_140173787 | 0.26 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr13_-_94383040 | 0.26 |
ENSMUST00000153558.8
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr13_-_99027544 | 0.26 |
ENSMUST00000109399.9
|
Tnpo1
|
transportin 1 |
| chr15_+_4404965 | 0.26 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr2_-_52448552 | 0.26 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chrX_+_152281200 | 0.26 |
ENSMUST00000060714.10
|
Ubqln2
|
ubiquilin 2 |
| chr5_+_150119860 | 0.25 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr9_-_103182246 | 0.25 |
ENSMUST00000142540.2
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr1_+_179788675 | 0.25 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_-_57483175 | 0.24 |
ENSMUST00000029380.14
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr9_-_88364593 | 0.24 |
ENSMUST00000173801.8
ENSMUST00000069221.12 ENSMUST00000172508.2 |
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr2_-_32178034 | 0.24 |
ENSMUST00000183946.8
ENSMUST00000113400.3 ENSMUST00000050410.11 |
Swi5
|
SWI5 recombination repair homolog (yeast) |
| chr5_+_117919082 | 0.24 |
ENSMUST00000138579.3
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr8_+_47193275 | 0.24 |
ENSMUST00000207571.3
|
Irf2
|
interferon regulatory factor 2 |
| chr7_-_84328553 | 0.24 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr14_+_53180476 | 0.24 |
ENSMUST00000103596.3
|
Trav13d-2
|
T cell receptor alpha variable 13D-2 |
| chr3_+_60436570 | 0.24 |
ENSMUST00000192607.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr14_+_53180221 | 0.23 |
ENSMUST00000197954.2
|
Trav13d-2
|
T cell receptor alpha variable 13D-2 |
| chr19_-_12742811 | 0.23 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr10_+_36383008 | 0.23 |
ENSMUST00000168572.8
|
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr11_-_54751738 | 0.23 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chrX_+_73468140 | 0.23 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr13_+_113931155 | 0.23 |
ENSMUST00000224068.2
|
Arl15
|
ADP-ribosylation factor-like 15 |
| chr1_+_127234441 | 0.23 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr19_-_40502117 | 0.23 |
ENSMUST00000225148.2
ENSMUST00000224667.2 ENSMUST00000225153.2 ENSMUST00000238831.2 ENSMUST00000226047.2 ENSMUST00000224247.2 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr11_-_78277384 | 0.23 |
ENSMUST00000108294.2
|
Foxn1
|
forkhead box N1 |
| chr11_-_102446947 | 0.23 |
ENSMUST00000143842.2
|
Gpatch8
|
G patch domain containing 8 |
| chr9_+_6168601 | 0.23 |
ENSMUST00000168039.8
|
Pdgfd
|
platelet-derived growth factor, D polypeptide |
| chr2_+_70393782 | 0.23 |
ENSMUST00000123330.3
|
Gad1
|
glutamate decarboxylase 1 |
| chr8_-_41469332 | 0.23 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr14_+_53491504 | 0.22 |
ENSMUST00000103622.3
|
Trav13n-2
|
T cell receptor alpha variable 13N-2 |
| chr2_+_105505823 | 0.22 |
ENSMUST00000167211.9
ENSMUST00000111083.10 |
Pax6
|
paired box 6 |
| chr14_+_32321341 | 0.22 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chrX_-_43879055 | 0.22 |
ENSMUST00000060481.9
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr4_-_56741398 | 0.22 |
ENSMUST00000095080.5
|
Actl7b
|
actin-like 7b |
| chr12_+_71062733 | 0.22 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_-_16761790 | 0.21 |
ENSMUST00000003183.12
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr9_+_32135540 | 0.21 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr10_+_90412638 | 0.21 |
ENSMUST00000183136.8
ENSMUST00000182595.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_+_25910510 | 0.21 |
ENSMUST00000228600.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr6_+_15185399 | 0.21 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr18_-_84104507 | 0.21 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr14_+_53995108 | 0.21 |
ENSMUST00000184905.2
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr16_+_43184191 | 0.21 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_+_93096316 | 0.20 |
ENSMUST00000138595.3
|
Crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr2_+_79538124 | 0.20 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr12_-_84240781 | 0.20 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr3_+_10431961 | 0.20 |
ENSMUST00000029049.7
|
Chmp4c
|
charged multivesicular body protein 4C |
| chrX_+_36390430 | 0.20 |
ENSMUST00000016553.5
|
Nkap
|
NFKB activating protein |
| chr16_+_20551853 | 0.20 |
ENSMUST00000115423.8
ENSMUST00000007171.13 ENSMUST00000232646.2 |
Chrd
|
chordin |
| chr3_+_87458556 | 0.20 |
ENSMUST00000238853.2
|
Etv3l
|
ets variant 3-like |
| chr14_+_53491249 | 0.19 |
ENSMUST00000196941.2
|
Trav13n-2
|
T cell receptor alpha variable 13N-2 |
| chr18_+_37108994 | 0.19 |
ENSMUST00000193984.2
|
Gm37388
|
predicted gene, 37388 |
| chr5_+_143349645 | 0.19 |
ENSMUST00000010969.15
|
Grid2ip
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein 1 |
| chr10_-_30531768 | 0.19 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr8_+_46081213 | 0.19 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_+_20151406 | 0.19 |
ENSMUST00000020358.12
ENSMUST00000109602.8 ENSMUST00000109601.8 ENSMUST00000163483.2 |
Rab1a
|
RAB1A, member RAS oncogene family |
| chr5_-_90487583 | 0.19 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr18_-_84104574 | 0.19 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr9_+_21279802 | 0.19 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr14_+_53399856 | 0.19 |
ENSMUST00000198359.2
|
Trav13n-1
|
T cell receptor alpha variable 13N-1 |
| chr16_-_34334314 | 0.19 |
ENSMUST00000151491.8
ENSMUST00000114960.9 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr15_+_79784543 | 0.19 |
ENSMUST00000230741.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr7_-_144761806 | 0.18 |
ENSMUST00000208788.2
|
Smim38
|
small integral membrane protein 38 |
| chr19_+_56450085 | 0.18 |
ENSMUST00000225909.2
|
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr16_+_43330630 | 0.18 |
ENSMUST00000114695.3
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_+_71511642 | 0.18 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr4_+_150233362 | 0.18 |
ENSMUST00000059893.8
|
Slc2a7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chr7_-_63586049 | 0.18 |
ENSMUST00000185175.2
|
Klf13
|
Kruppel-like factor 13 |
| chr2_-_45000250 | 0.18 |
ENSMUST00000201211.4
ENSMUST00000177302.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_102231208 | 0.18 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr19_+_56450062 | 0.18 |
ENSMUST00000178590.9
ENSMUST00000039666.8 |
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr9_-_54408780 | 0.18 |
ENSMUST00000118600.8
ENSMUST00000118163.8 |
Dmxl2
|
Dmx-like 2 |
| chr3_-_85648696 | 0.18 |
ENSMUST00000094148.6
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr2_-_63928339 | 0.18 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr14_+_53921052 | 0.18 |
ENSMUST00000103663.6
|
Trav4-4-dv10
|
T cell receptor alpha variable 4-4-DV10 |
| chr10_+_90412570 | 0.17 |
ENSMUST00000182430.8
ENSMUST00000182960.8 ENSMUST00000182045.2 ENSMUST00000182083.2 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr13_+_41071077 | 0.17 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr10_-_30531832 | 0.17 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr8_+_47192911 | 0.17 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr4_+_123077515 | 0.17 |
ENSMUST00000152194.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr14_+_51441146 | 0.17 |
ENSMUST00000049559.2
|
Ear14
|
eosinophil-associated, ribonuclease A family, member 14 |
| chr16_+_39804711 | 0.17 |
ENSMUST00000187695.7
|
Lsamp
|
limbic system-associated membrane protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.5 | 1.9 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.4 | 1.1 | GO:2000184 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.3 | 2.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 1.7 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 1.3 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.2 | 0.5 | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000299) |
| 0.2 | 1.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 1.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.2 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 | 1.8 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 1.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.3 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.1 | 0.4 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.4 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.3 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.3 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.2 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.2 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 0.4 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 0.6 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.7 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 1.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 3.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.4 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.5 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.3 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 0.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.4 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.7 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.2 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.2 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 1.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0099545 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:0060128 | dorsal/ventral axon guidance(GO:0033563) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.1 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.9 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.1 | GO:0032430 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.0 | 0.1 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.4 | 1.1 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.1 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0097452 | CRD-mediated mRNA stability complex(GO:0070937) GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.2 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.6 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.2 | 1.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 2.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 0.7 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 0.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 3.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.4 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.1 | 1.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.6 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.3 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 1.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0018455 | alcohol dehydrogenase [NAD(P)+] activity(GO:0018455) |
| 0.0 | 0.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 1.3 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.5 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.7 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |