Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
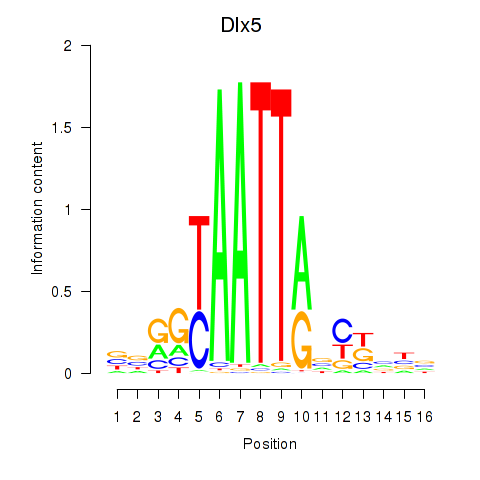
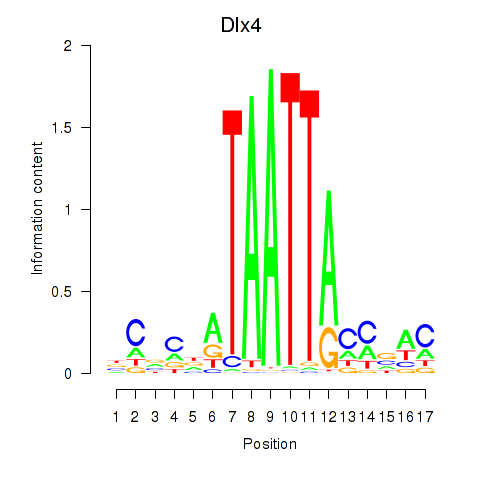
Results for Dlx5_Dlx4
Z-value: 0.95
Transcription factors associated with Dlx5_Dlx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dlx5
|
ENSMUSG00000029755.11 | Dlx5 |
|
Dlx4
|
ENSMUSG00000020871.9 | Dlx4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx5 | mm39_v1_chr6_-_6882068_6882092 | 0.43 | 9.4e-03 | Click! |
| Dlx4 | mm39_v1_chr11_-_95037089_95037089 | -0.00 | 9.9e-01 | Click! |
Activity profile of Dlx5_Dlx4 motif
Sorted Z-values of Dlx5_Dlx4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Dlx5_Dlx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41121979 | 7.10 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr11_+_87684299 | 5.46 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr9_-_21671571 | 3.24 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr11_-_11920540 | 3.23 |
ENSMUST00000109653.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chrX_+_149330371 | 3.14 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr19_-_40576782 | 3.08 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr8_-_4829519 | 2.99 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr3_-_106126794 | 2.74 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr19_-_40576817 | 2.72 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr3_-_98247237 | 2.65 |
ENSMUST00000065793.12
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr4_-_131802561 | 2.38 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr10_-_75946790 | 2.33 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr12_-_75678092 | 2.32 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr7_+_100145192 | 2.27 |
ENSMUST00000133044.3
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr16_-_75706161 | 2.26 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr4_-_131802606 | 2.23 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr8_+_84682136 | 2.22 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chrX_+_139857640 | 2.08 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr11_-_117671436 | 2.02 |
ENSMUST00000026659.10
ENSMUST00000127227.2 |
Tmc6
|
transmembrane channel-like gene family 6 |
| chr13_+_23930717 | 2.00 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr9_+_65797519 | 1.98 |
ENSMUST00000045802.7
|
Pclaf
|
PCNA clamp associated factor |
| chr8_+_117648474 | 1.98 |
ENSMUST00000034205.5
ENSMUST00000212775.2 |
Cenpn
|
centromere protein N |
| chr14_+_62529924 | 1.96 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chrX_+_139857688 | 1.96 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr7_-_133304244 | 1.93 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chr1_-_173161069 | 1.91 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr6_+_72074545 | 1.88 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr7_+_126376099 | 1.78 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr2_+_118644717 | 1.77 |
ENSMUST00000028803.14
ENSMUST00000126045.8 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr9_-_20871081 | 1.76 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr4_-_43499608 | 1.74 |
ENSMUST00000136005.3
ENSMUST00000054538.13 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr8_+_106024294 | 1.71 |
ENSMUST00000015003.10
|
E2f4
|
E2F transcription factor 4 |
| chr2_+_174292471 | 1.68 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr2_-_32277773 | 1.68 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr1_+_152683627 | 1.64 |
ENSMUST00000027754.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr17_-_36149142 | 1.56 |
ENSMUST00000001566.10
|
Tubb5
|
tubulin, beta 5 class I |
| chr2_+_118644675 | 1.56 |
ENSMUST00000110842.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr3_+_103078971 | 1.53 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr2_-_32278245 | 1.53 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr8_-_65489834 | 1.53 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr11_-_16958647 | 1.52 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr7_-_103778992 | 1.52 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr8_-_86091946 | 1.49 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr13_+_51799268 | 1.48 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr2_-_131001916 | 1.46 |
ENSMUST00000103188.10
ENSMUST00000133602.8 ENSMUST00000028800.12 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr12_-_55061117 | 1.45 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr17_-_36149100 | 1.42 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr10_+_115653152 | 1.41 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr10_+_20223516 | 1.41 |
ENSMUST00000169712.3
ENSMUST00000217608.2 |
Mtfr2
|
mitochondrial fission regulator 2 |
| chr10_+_58159288 | 1.40 |
ENSMUST00000020078.14
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_+_118644475 | 1.40 |
ENSMUST00000134661.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chrX_+_55500170 | 1.30 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr11_+_58839716 | 1.28 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chrX_+_92718695 | 1.26 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr2_-_85966272 | 1.22 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr16_+_45430743 | 1.22 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr11_-_26543915 | 1.21 |
ENSMUST00000078362.13
|
Vrk2
|
vaccinia related kinase 2 |
| chr14_-_67953035 | 1.20 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr18_+_44237474 | 1.20 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr2_+_110427643 | 1.18 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr4_-_156340713 | 1.16 |
ENSMUST00000219393.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr9_-_58648826 | 1.14 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr2_+_125089110 | 1.12 |
ENSMUST00000082122.14
|
Dut
|
deoxyuridine triphosphatase |
| chr6_-_56900917 | 1.12 |
ENSMUST00000031793.8
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr11_-_53371050 | 1.11 |
ENSMUST00000104955.4
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr10_+_75399920 | 1.10 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_+_82817794 | 1.09 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr6_+_5390386 | 1.08 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr5_-_116162415 | 1.06 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr17_+_47922497 | 1.05 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr8_-_65489791 | 1.03 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chrX_+_133486391 | 1.03 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr4_-_41464816 | 1.01 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr10_-_62363217 | 1.01 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chr4_+_19818718 | 1.00 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr14_+_26722319 | 1.00 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chrX_+_164953444 | 0.98 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr10_-_129738595 | 0.96 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr2_+_91376650 | 0.95 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr1_+_40478787 | 0.93 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr17_-_47922374 | 0.93 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr6_-_56900709 | 0.93 |
ENSMUST00000205087.2
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr17_-_33937565 | 0.93 |
ENSMUST00000174040.2
ENSMUST00000173015.8 ENSMUST00000066121.13 ENSMUST00000186022.7 ENSMUST00000173329.8 ENSMUST00000172767.9 |
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr3_+_93427791 | 0.93 |
ENSMUST00000029515.5
|
S100a11
|
S100 calcium binding protein A11 |
| chr9_-_36637670 | 0.90 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr2_+_87610895 | 0.90 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152 |
| chr17_+_21060706 | 0.90 |
ENSMUST00000232909.2
ENSMUST00000233670.2 ENSMUST00000233939.2 |
Vmn1r230
|
vomeronasal 1 receptor 230 |
| chr6_-_83010402 | 0.90 |
ENSMUST00000089651.6
|
Dok1
|
docking protein 1 |
| chr7_-_103094646 | 0.88 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr19_-_6065872 | 0.88 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1 |
| chr13_+_21900554 | 0.87 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr1_+_160898283 | 0.87 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr11_-_26543970 | 0.86 |
ENSMUST00000156264.2
|
Vrk2
|
vaccinia related kinase 2 |
| chr9_-_62719208 | 0.85 |
ENSMUST00000034775.10
|
Fem1b
|
fem 1 homolog b |
| chr12_+_105996961 | 0.84 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr4_-_117013396 | 0.83 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
| chrX_+_158086253 | 0.83 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr7_+_108265625 | 0.83 |
ENSMUST00000213979.3
ENSMUST00000216331.2 ENSMUST00000217170.2 |
Olfr510
|
olfactory receptor 510 |
| chr17_-_36331416 | 0.83 |
ENSMUST00000174063.2
ENSMUST00000113760.10 |
H2-T24
|
histocompatibility 2, T region locus 24 |
| chr8_-_117648147 | 0.82 |
ENSMUST00000078589.7
ENSMUST00000148235.8 |
Cmc2
|
COX assembly mitochondrial protein 2 |
| chr18_+_44237577 | 0.80 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr5_-_129864202 | 0.80 |
ENSMUST00000136507.4
|
Psph
|
phosphoserine phosphatase |
| chr5_-_65855511 | 0.80 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr9_-_36637923 | 0.79 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr1_+_85538554 | 0.79 |
ENSMUST00000162925.2
|
Sp140
|
Sp140 nuclear body protein |
| chr11_-_73382303 | 0.78 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr9_-_44024767 | 0.77 |
ENSMUST00000216511.2
ENSMUST00000056328.6 ENSMUST00000185479.2 |
Rnf26
Gm49380
|
ring finger protein 26 predicted gene, 49380 |
| chr4_+_126450728 | 0.77 |
ENSMUST00000048391.15
|
Clspn
|
claspin |
| chr5_-_137531471 | 0.77 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr6_-_87815653 | 0.76 |
ENSMUST00000204431.2
ENSMUST00000089497.7 |
Isy1
|
ISY1 splicing factor homolog |
| chr9_-_111086528 | 0.74 |
ENSMUST00000199404.2
|
Mlh1
|
mutL homolog 1 |
| chr5_-_137531413 | 0.74 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr11_+_52265090 | 0.73 |
ENSMUST00000020673.3
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr6_-_66537080 | 0.73 |
ENSMUST00000079584.3
ENSMUST00000227014.2 |
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr10_-_111833138 | 0.73 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr13_+_35843816 | 0.73 |
ENSMUST00000075220.14
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr5_-_138185438 | 0.72 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr12_-_40087393 | 0.72 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr15_-_78947038 | 0.72 |
ENSMUST00000151889.8
ENSMUST00000040676.11 |
Ankrd54
|
ankyrin repeat domain 54 |
| chr2_-_86109346 | 0.72 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr14_-_47514248 | 0.71 |
ENSMUST00000187531.8
ENSMUST00000111790.2 |
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr14_-_52341426 | 0.71 |
ENSMUST00000227536.2
ENSMUST00000227195.2 ENSMUST00000228815.2 ENSMUST00000228198.2 ENSMUST00000227458.2 ENSMUST00000228232.2 ENSMUST00000227242.2 ENSMUST00000228748.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr5_-_138169476 | 0.70 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr14_-_52341472 | 0.70 |
ENSMUST00000111610.12
ENSMUST00000164655.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr8_+_85415935 | 0.70 |
ENSMUST00000125370.10
ENSMUST00000175784.2 |
Trmt1
|
tRNA methyltransferase 1 |
| chr5_-_137531463 | 0.70 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr2_+_155593030 | 0.70 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr8_-_70963202 | 0.68 |
ENSMUST00000125184.8
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr7_+_101545547 | 0.68 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr17_+_46807637 | 0.68 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr9_+_19620729 | 0.67 |
ENSMUST00000217450.4
ENSMUST00000212013.4 |
Olfr857
|
olfactory receptor 857 |
| chr14_+_19801333 | 0.67 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr11_-_30936326 | 0.67 |
ENSMUST00000020553.5
ENSMUST00000101394.5 |
Chac2
|
ChaC, cation transport regulator 2 |
| chr2_+_85838122 | 0.67 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
| chr2_-_126342551 | 0.66 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr19_+_13208692 | 0.65 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr9_-_103357564 | 0.65 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr6_-_122317484 | 0.64 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr13_+_21901791 | 0.64 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr9_+_19716202 | 0.63 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr7_-_44578834 | 0.63 |
ENSMUST00000107857.11
ENSMUST00000167930.8 ENSMUST00000085399.13 ENSMUST00000166972.9 |
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr5_+_145020640 | 0.62 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr10_+_129219952 | 0.62 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr19_-_8775935 | 0.62 |
ENSMUST00000096261.5
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chrX_-_52328963 | 0.62 |
ENSMUST00000074861.9
|
Plac1
|
placental specific protein 1 |
| chr7_-_30523191 | 0.62 |
ENSMUST00000053156.10
|
Ffar2
|
free fatty acid receptor 2 |
| chr11_-_58059293 | 0.62 |
ENSMUST00000172035.8
ENSMUST00000035604.13 ENSMUST00000102711.9 |
Gemin5
|
gem nuclear organelle associated protein 5 |
| chr8_-_117648104 | 0.61 |
ENSMUST00000128304.2
|
Cmc2
|
COX assembly mitochondrial protein 2 |
| chr11_-_69786324 | 0.60 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr1_+_40478926 | 0.60 |
ENSMUST00000173514.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr9_-_109455125 | 0.59 |
ENSMUST00000073962.8
|
Fbxw24
|
F-box and WD-40 domain protein 24 |
| chr12_+_55171254 | 0.59 |
ENSMUST00000177768.3
|
Fam177a
|
family with sequence similarity 177, member A |
| chr19_+_12647803 | 0.59 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr9_+_38164070 | 0.59 |
ENSMUST00000213129.2
|
Olfr143
|
olfactory receptor 143 |
| chr18_+_69652751 | 0.58 |
ENSMUST00000200966.4
|
Tcf4
|
transcription factor 4 |
| chr11_-_117670430 | 0.58 |
ENSMUST00000143406.8
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr2_-_86528739 | 0.58 |
ENSMUST00000214141.2
|
Olfr1087
|
olfactory receptor 1087 |
| chr19_+_34078333 | 0.58 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr2_-_151586063 | 0.57 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr19_+_55169148 | 0.57 |
ENSMUST00000154886.8
ENSMUST00000120936.8 ENSMUST00000025936.12 |
Tectb
|
tectorin beta |
| chr1_+_139382485 | 0.57 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr2_+_36120438 | 0.57 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr19_-_8775817 | 0.57 |
ENSMUST00000235964.2
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr10_-_23977810 | 0.57 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr19_-_6065799 | 0.56 |
ENSMUST00000235138.2
|
Capn1
|
calpain 1 |
| chr3_+_103739877 | 0.56 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr2_-_111965322 | 0.56 |
ENSMUST00000213696.2
|
Olfr1316
|
olfactory receptor 1316 |
| chr8_+_23901506 | 0.56 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr5_-_138185686 | 0.55 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr5_+_4073343 | 0.55 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr10_-_128361731 | 0.55 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr18_+_69652550 | 0.55 |
ENSMUST00000201205.4
|
Tcf4
|
transcription factor 4 |
| chr3_-_72875187 | 0.55 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr8_+_3410842 | 0.55 |
ENSMUST00000238813.2
|
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr1_+_131838294 | 0.54 |
ENSMUST00000062264.8
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr7_+_108533613 | 0.54 |
ENSMUST00000033342.7
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr7_-_30259253 | 0.54 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr8_-_22550035 | 0.54 |
ENSMUST00000110738.3
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chrX_+_105626790 | 0.54 |
ENSMUST00000101296.9
ENSMUST00000101297.4 |
Gm5127
|
predicted gene 5127 |
| chr9_+_123195986 | 0.54 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr11_-_109502243 | 0.53 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr6_-_57827328 | 0.53 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr17_+_66418525 | 0.53 |
ENSMUST00000072383.14
|
Washc1
|
WASH complex subunit 1 |
| chrX_-_133012600 | 0.53 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr2_+_87853118 | 0.53 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr18_+_69652837 | 0.52 |
ENSMUST00000201410.4
ENSMUST00000202937.4 |
Tcf4
|
transcription factor 4 |
| chr8_-_120331936 | 0.52 |
ENSMUST00000093099.13
|
Taf1c
|
TATA-box binding protein associated factor, RNA polymerase I, C |
| chr7_+_101546059 | 0.52 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr4_-_59783780 | 0.52 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr14_-_104760051 | 0.52 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr11_+_87000032 | 0.52 |
ENSMUST00000020794.6
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr7_+_126376353 | 0.52 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr17_+_28059099 | 0.51 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr17_+_28059129 | 0.51 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr13_-_12272962 | 0.51 |
ENSMUST00000099856.6
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr2_+_150751475 | 0.51 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr14_+_32043944 | 0.51 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chrX_-_8118541 | 0.50 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr3_-_116047148 | 0.50 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr17_+_29312737 | 0.50 |
ENSMUST00000023829.8
ENSMUST00000233296.2 |
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.8 | 5.5 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.8 | 7.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.1 | 3.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.6 | 1.9 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.6 | 4.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.6 | 1.7 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.5 | 2.6 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.5 | 1.5 | GO:0060305 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.5 | 1.4 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.4 | 1.8 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 | 1.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.3 | 3.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.3 | 2.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 1.1 | GO:0009223 | dUMP biosynthetic process(GO:0006226) pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.3 | 1.1 | GO:0033380 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.3 | 4.6 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 0.8 | GO:0000389 | generation of catalytic spliceosome for second transesterification step(GO:0000350) mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 1.5 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 0.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 0.9 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 2.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.2 | 2.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 4.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 0.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 3.9 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 0.8 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 0.8 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.2 | 0.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 2.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.8 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 0.6 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 0.7 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 0.7 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.2 | 0.5 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.2 | 0.5 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.2 | 0.5 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.2 | 1.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 2.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 0.5 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 0.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.8 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 0.6 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.2 | 0.5 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.2 | 1.8 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 0.7 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 1.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 1.1 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.7 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.4 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.1 | 2.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.5 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.5 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.5 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 1.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 2.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.3 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.6 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 1.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 2.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.6 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 2.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 0.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 1.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.6 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 2.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.2 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.4 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.3 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 | 0.2 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.1 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.4 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.1 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.1 | 0.2 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.5 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 1.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 2.7 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.3 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.4 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.2 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.1 | 2.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.5 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 1.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.2 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 0.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 2.0 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.3 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.3 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.7 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0009814 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.6 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 4.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:1904109 | positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 4.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 32.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 2.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.4 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.6 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.5 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 2.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.3 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.5 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.3 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.5 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.3 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.9 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.8 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0021995 | optic cup structural organization(GO:0003409) anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.2 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.0 | 1.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 2.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 1.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 1.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 1.0 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 1.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 1.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.0 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.4 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.7 | GO:0071300 | histone ubiquitination(GO:0016574) cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.6 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.5 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 1.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 3.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 2.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 5.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 0.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 0.7 | GO:0005715 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.2 | 2.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 2.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 1.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 6.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.6 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 1.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 0.5 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 1.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 4.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.1 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 0.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 3.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 3.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 0.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.0 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0097422 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 2.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 4.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0019202 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 1.0 | 3.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.6 | 1.9 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.5 | 1.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.4 | 2.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 7.1 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.3 | 1.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 1.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 1.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.3 | 2.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 0.8 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.3 | 2.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 0.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 1.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.3 | 1.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 1.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 2.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.6 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.2 | 2.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.8 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 0.7 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.2 | 0.5 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.2 | 0.5 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.2 | 1.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.5 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 1.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 3.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.5 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 2.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.6 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 3.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 2.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 6.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.3 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.4 | GO:0016623 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.4 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 18.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.3 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 4.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 5.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.4 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 3.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 2.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 21.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.8 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.5 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0051381 | histamine binding(GO:0051381) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:1902121 | NADP+ binding(GO:0070401) lithocholic acid binding(GO:1902121) |
| 0.0 | 0.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.8 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0015450 | protein channel activity(GO:0015266) P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 5.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 5.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 3.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 3.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 5.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 7.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 4.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 6.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 2.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 1.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 2.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |