Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Ebf3
Z-value: 3.93
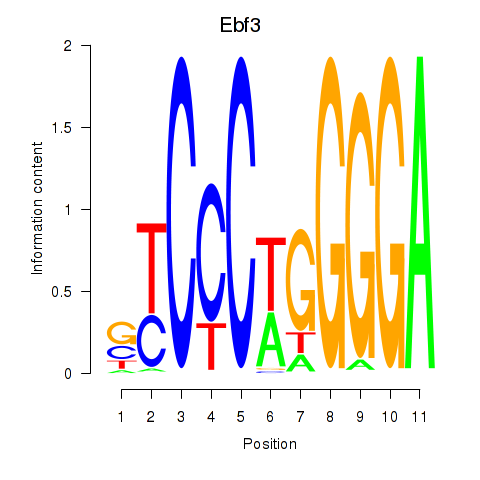
Transcription factors associated with Ebf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ebf3
|
ENSMUSG00000010476.15 | Ebf3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ebf3 | mm39_v1_chr7_-_136916123_136916174 | 0.43 | 9.1e-03 | Click! |
Activity profile of Ebf3 motif
Sorted Z-values of Ebf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ebf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102255999 | 41.83 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chrX_-_7834057 | 32.58 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr2_-_28511941 | 25.83 |
ENSMUST00000028156.8
ENSMUST00000164290.8 |
Gfi1b
|
growth factor independent 1B |
| chr12_+_109419371 | 25.22 |
ENSMUST00000109844.11
ENSMUST00000109842.9 ENSMUST00000109843.8 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr17_-_48758538 | 22.34 |
ENSMUST00000024794.12
|
Tspo2
|
translocator protein 2 |
| chr14_-_69740670 | 19.77 |
ENSMUST00000180059.3
ENSMUST00000179116.3 |
Gm16867
|
predicted gene, 16867 |
| chr12_+_109419575 | 19.64 |
ENSMUST00000173539.8
ENSMUST00000109841.9 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr10_-_128237087 | 18.58 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr11_+_94901104 | 16.52 |
ENSMUST00000124735.2
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr2_-_27974889 | 15.20 |
ENSMUST00000028179.15
ENSMUST00000117486.8 ENSMUST00000135472.2 |
Fcnb
|
ficolin B |
| chr7_+_24596806 | 13.67 |
ENSMUST00000003469.8
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr12_+_109419454 | 13.46 |
ENSMUST00000109846.11
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr14_-_70866385 | 13.00 |
ENSMUST00000228824.2
|
Dmtn
|
dematin actin binding protein |
| chr17_+_48666919 | 12.81 |
ENSMUST00000224001.2
ENSMUST00000024792.8 ENSMUST00000225849.2 |
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr14_-_70864666 | 12.23 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chrX_+_47235313 | 11.09 |
ENSMUST00000033427.7
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr7_-_126014027 | 10.83 |
ENSMUST00000032968.7
ENSMUST00000206325.2 |
Cd19
|
CD19 antigen |
| chr4_+_120523758 | 9.97 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr1_-_171108754 | 9.81 |
ENSMUST00000073120.11
|
Ppox
|
protoporphyrinogen oxidase |
| chr7_+_28682253 | 9.70 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr6_-_86646118 | 9.26 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr19_+_58717319 | 9.25 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr15_+_78128990 | 9.01 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr9_+_95519654 | 8.92 |
ENSMUST00000015498.9
|
Pcolce2
|
procollagen C-endopeptidase enhancer 2 |
| chr6_+_30639217 | 8.66 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chrX_+_74425990 | 8.60 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr11_+_115768323 | 8.49 |
ENSMUST00000222123.2
|
Myo15b
|
myosin XVB |
| chr15_+_80507671 | 8.36 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr7_+_141995545 | 8.22 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr15_-_66684442 | 8.11 |
ENSMUST00000100572.10
|
Sla
|
src-like adaptor |
| chr14_-_60324265 | 7.76 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr11_-_53371050 | 7.58 |
ENSMUST00000104955.4
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr7_+_141996067 | 7.43 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr11_-_11758923 | 7.28 |
ENSMUST00000109664.2
ENSMUST00000150714.2 ENSMUST00000047689.11 ENSMUST00000171080.8 ENSMUST00000171938.2 |
Fignl1
|
fidgetin-like 1 |
| chr7_-_24997291 | 7.28 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr11_+_115790951 | 7.19 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr11_+_101207743 | 7.07 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr4_+_43957401 | 6.91 |
ENSMUST00000030202.14
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr11_+_63019799 | 6.89 |
ENSMUST00000108702.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr15_+_78129040 | 6.84 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr10_+_60182630 | 6.71 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr17_-_7050145 | 6.69 |
ENSMUST00000064234.7
|
Ezr
|
ezrin |
| chr8_+_85696695 | 6.68 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr5_+_137627431 | 6.63 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr7_+_79836581 | 6.45 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr10_+_41352310 | 6.35 |
ENSMUST00000019967.16
ENSMUST00000119962.8 ENSMUST00000099934.11 |
Mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr5_+_136996713 | 6.32 |
ENSMUST00000001790.6
|
Cldn15
|
claudin 15 |
| chr9_-_58648826 | 6.31 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr12_-_113542610 | 6.31 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr15_+_79400597 | 6.06 |
ENSMUST00000010974.9
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chrX_-_161747552 | 6.00 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr4_+_43957677 | 5.98 |
ENSMUST00000107855.2
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr9_+_103182352 | 5.92 |
ENSMUST00000035164.10
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr16_+_32427738 | 5.92 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr2_+_25262327 | 5.88 |
ENSMUST00000028329.13
ENSMUST00000114293.9 ENSMUST00000100323.3 |
Sapcd2
|
suppressor APC domain containing 2 |
| chr18_-_35781422 | 5.85 |
ENSMUST00000237462.2
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr11_+_70323452 | 5.80 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr7_-_45570254 | 5.79 |
ENSMUST00000164119.3
|
Emp3
|
epithelial membrane protein 3 |
| chr7_+_127661835 | 5.73 |
ENSMUST00000106242.10
ENSMUST00000120355.8 ENSMUST00000106240.9 ENSMUST00000098015.10 |
Itgam
Gm49368
|
integrin alpha M predicted gene, 49368 |
| chr12_-_113823290 | 5.70 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr16_+_32427789 | 5.64 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr7_+_44465353 | 5.61 |
ENSMUST00000208626.2
ENSMUST00000057195.17 |
Nup62
|
nucleoporin 62 |
| chr2_+_30331839 | 5.58 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr9_+_45314436 | 5.45 |
ENSMUST00000041005.6
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr11_+_101207021 | 5.36 |
ENSMUST00000142640.8
ENSMUST00000019470.14 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr6_-_91093766 | 5.29 |
ENSMUST00000113509.2
ENSMUST00000032179.14 |
Nup210
|
nucleoporin 210 |
| chr11_-_3489228 | 5.18 |
ENSMUST00000075118.10
ENSMUST00000136243.2 ENSMUST00000020721.15 ENSMUST00000170588.8 |
Smtn
|
smoothelin |
| chr12_-_113561594 | 5.07 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr4_+_130643260 | 5.04 |
ENSMUST00000030316.7
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr1_+_75376714 | 5.02 |
ENSMUST00000113589.8
|
Speg
|
SPEG complex locus |
| chr11_+_75541324 | 4.93 |
ENSMUST00000102505.10
|
Myo1c
|
myosin IC |
| chr7_-_3680530 | 4.88 |
ENSMUST00000038743.15
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr1_+_74414354 | 4.84 |
ENSMUST00000187516.7
ENSMUST00000027368.6 |
Slc11a1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
| chr13_+_55547498 | 4.84 |
ENSMUST00000057167.9
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr7_-_115630282 | 4.76 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr17_-_80514725 | 4.73 |
ENSMUST00000234696.2
ENSMUST00000235069.2 ENSMUST00000063417.11 |
Srsf7
|
serine and arginine-rich splicing factor 7 |
| chr11_+_96820091 | 4.65 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr11_+_3939924 | 4.60 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr1_+_172328768 | 4.59 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr4_+_140875222 | 4.45 |
ENSMUST00000030757.10
|
Fbxo42
|
F-box protein 42 |
| chr17_+_36176485 | 4.40 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr7_+_19144950 | 4.38 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chrX_+_10351360 | 4.37 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr1_+_82817794 | 4.25 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr11_+_96820220 | 4.15 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr2_-_153083322 | 4.14 |
ENSMUST00000056924.14
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr7_-_142253247 | 4.12 |
ENSMUST00000105934.8
|
Ins2
|
insulin II |
| chr12_-_113860566 | 4.07 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr11_-_116001037 | 4.03 |
ENSMUST00000106441.8
ENSMUST00000021120.6 |
Trim47
|
tripartite motif-containing 47 |
| chr17_+_47983587 | 4.00 |
ENSMUST00000152724.2
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr14_+_62529924 | 3.97 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr2_+_154390808 | 3.93 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr1_-_136161850 | 3.91 |
ENSMUST00000120339.8
|
Inava
|
innate immunity activator |
| chr2_+_124910037 | 3.89 |
ENSMUST00000070353.4
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr15_+_85220438 | 3.88 |
ENSMUST00000163242.3
|
Atxn10
|
ataxin 10 |
| chr12_-_113589576 | 3.86 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr8_+_108271643 | 3.85 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr19_-_39875192 | 3.84 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr2_-_126341757 | 3.72 |
ENSMUST00000040128.12
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr14_+_62569517 | 3.71 |
ENSMUST00000022499.13
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr6_-_116650751 | 3.70 |
ENSMUST00000204576.2
ENSMUST00000203029.3 ENSMUST00000035842.7 |
Rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr17_+_84013575 | 3.65 |
ENSMUST00000112350.8
ENSMUST00000112349.9 ENSMUST00000112352.10 ENSMUST00000067826.15 |
Mta3
|
metastasis associated 3 |
| chr4_+_117692583 | 3.64 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr12_-_113700190 | 3.61 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr11_-_69728560 | 3.60 |
ENSMUST00000108634.9
|
Nlgn2
|
neuroligin 2 |
| chr5_+_137627376 | 3.59 |
ENSMUST00000031734.16
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr6_+_67586695 | 3.58 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr18_+_34757687 | 3.56 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chr7_-_44741622 | 3.54 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr9_+_108356935 | 3.43 |
ENSMUST00000194147.2
ENSMUST00000065014.10 ENSMUST00000195483.6 ENSMUST00000195058.2 |
Lamb2
|
laminin, beta 2 |
| chr15_+_99600149 | 3.38 |
ENSMUST00000229236.2
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr11_-_102208449 | 3.36 |
ENSMUST00000107123.10
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr4_+_137408975 | 3.29 |
ENSMUST00000047243.12
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr18_+_34757666 | 3.24 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr7_+_127661807 | 3.16 |
ENSMUST00000064821.14
|
Itgam
|
integrin alpha M |
| chr1_+_172139934 | 3.13 |
ENSMUST00000039506.15
|
Igsf8
|
immunoglobulin superfamily, member 8 |
| chr2_-_13496624 | 3.12 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr6_+_68279392 | 3.08 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr4_+_128582519 | 3.01 |
ENSMUST00000106080.8
|
Phc2
|
polyhomeotic 2 |
| chr9_-_58220469 | 3.01 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr8_-_72154405 | 2.95 |
ENSMUST00000034260.9
|
B3gnt3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr12_-_113666198 | 2.94 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr17_-_46464441 | 2.92 |
ENSMUST00000171172.3
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr7_-_44741609 | 2.92 |
ENSMUST00000210734.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr15_+_78798116 | 2.89 |
ENSMUST00000089378.5
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chrX_+_133657312 | 2.88 |
ENSMUST00000081834.10
ENSMUST00000086880.11 ENSMUST00000086884.5 |
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr4_-_137523659 | 2.87 |
ENSMUST00000030551.11
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr11_+_101137786 | 2.86 |
ENSMUST00000107282.4
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr1_-_165462020 | 2.85 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chr4_-_149760488 | 2.84 |
ENSMUST00000118704.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr7_-_28071658 | 2.83 |
ENSMUST00000094644.11
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr2_+_130137068 | 2.80 |
ENSMUST00000110288.9
|
Ebf4
|
early B cell factor 4 |
| chr7_-_126224848 | 2.79 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr3_+_89979948 | 2.75 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr11_-_102207486 | 2.75 |
ENSMUST00000146896.9
ENSMUST00000079589.11 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr15_+_99599978 | 2.74 |
ENSMUST00000023759.6
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr9_-_66950991 | 2.72 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chr4_-_129436465 | 2.72 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr7_-_44465043 | 2.66 |
ENSMUST00000107893.9
|
Atf5
|
activating transcription factor 5 |
| chr8_+_71921824 | 2.66 |
ENSMUST00000124745.8
ENSMUST00000138892.2 ENSMUST00000147642.2 |
Dda1
|
DET1 and DDB1 associated 1 |
| chr17_+_36176948 | 2.58 |
ENSMUST00000122899.8
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_+_135768595 | 2.56 |
ENSMUST00000112087.9
ENSMUST00000178854.8 ENSMUST00000027671.12 ENSMUST00000179863.8 ENSMUST00000112085.9 ENSMUST00000112086.3 |
Tnnt2
|
troponin T2, cardiac |
| chr17_-_48235325 | 2.55 |
ENSMUST00000113263.8
ENSMUST00000097311.9 |
Foxp4
|
forkhead box P4 |
| chr11_-_96868483 | 2.55 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr11_-_102207516 | 2.54 |
ENSMUST00000107115.8
ENSMUST00000128016.2 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr9_-_44199428 | 2.54 |
ENSMUST00000160384.2
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chr10_-_87982732 | 2.53 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr2_-_34262012 | 2.50 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr11_+_76900091 | 2.46 |
ENSMUST00000129572.3
|
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr10_-_120037464 | 2.44 |
ENSMUST00000020448.11
|
Irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr5_-_18054702 | 2.44 |
ENSMUST00000165232.8
|
Cd36
|
CD36 molecule |
| chr19_-_45804446 | 2.43 |
ENSMUST00000079431.10
ENSMUST00000026247.13 ENSMUST00000162528.9 |
Kcnip2
|
Kv channel-interacting protein 2 |
| chr9_-_66956425 | 2.39 |
ENSMUST00000113687.8
ENSMUST00000113693.8 ENSMUST00000113701.8 ENSMUST00000034928.12 ENSMUST00000113685.10 ENSMUST00000030185.5 ENSMUST00000050905.16 ENSMUST00000113705.8 ENSMUST00000113697.8 ENSMUST00000113707.9 |
Tpm1
|
tropomyosin 1, alpha |
| chr6_-_69282389 | 2.37 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr5_+_65505657 | 2.35 |
ENSMUST00000031096.11
|
Klb
|
klotho beta |
| chr12_-_113912416 | 2.34 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr14_-_73563212 | 2.31 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chrX_-_100311824 | 2.30 |
ENSMUST00000033664.14
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr8_+_69333143 | 2.26 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr2_+_152596075 | 2.24 |
ENSMUST00000010020.12
|
Cox4i2
|
cytochrome c oxidase subunit 4I2 |
| chr15_+_99600475 | 2.24 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr7_-_126807581 | 2.23 |
ENSMUST00000120705.3
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr9_-_30833748 | 2.21 |
ENSMUST00000065112.7
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr12_-_114487525 | 2.15 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr5_-_147337162 | 2.10 |
ENSMUST00000049324.13
|
Flt3
|
FMS-like tyrosine kinase 3 |
| chr11_-_97944239 | 2.07 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr5_-_18054781 | 2.06 |
ENSMUST00000170051.8
|
Cd36
|
CD36 molecule |
| chr15_-_26895660 | 2.04 |
ENSMUST00000059204.11
|
Fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr2_+_32253692 | 2.03 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr11_-_115967873 | 2.00 |
ENSMUST00000153408.8
|
Unc13d
|
unc-13 homolog D |
| chr2_-_128529274 | 1.98 |
ENSMUST00000110332.2
ENSMUST00000110333.2 ENSMUST00000014499.10 |
Anapc1
|
anaphase promoting complex subunit 1 |
| chr16_+_32477722 | 1.96 |
ENSMUST00000238891.2
|
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr17_-_32607859 | 1.95 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr11_-_95733235 | 1.87 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr6_+_91450677 | 1.80 |
ENSMUST00000032183.6
|
Tmem43
|
transmembrane protein 43 |
| chr1_+_74317709 | 1.79 |
ENSMUST00000077985.4
|
Gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr14_-_70443471 | 1.79 |
ENSMUST00000227653.2
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr5_-_74229021 | 1.77 |
ENSMUST00000119154.8
ENSMUST00000068058.14 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr10_-_127587576 | 1.75 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr8_-_95644639 | 1.75 |
ENSMUST00000077955.6
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr9_-_66951025 | 1.75 |
ENSMUST00000113695.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr5_+_129802127 | 1.74 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr16_+_18210495 | 1.73 |
ENSMUST00000239548.1
|
ARVCF
|
armadillo repeat deleted in velocardiofacial syndrome |
| chr10_-_60055082 | 1.73 |
ENSMUST00000135158.9
|
Chst3
|
carbohydrate sulfotransferase 3 |
| chr19_-_10847121 | 1.71 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr7_-_100581314 | 1.70 |
ENSMUST00000107032.3
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr7_-_68398917 | 1.69 |
ENSMUST00000118110.3
|
Arrdc4
|
arrestin domain containing 4 |
| chr12_-_113790741 | 1.69 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr3_-_107667499 | 1.69 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr9_-_32454157 | 1.63 |
ENSMUST00000183767.2
|
Fli1
|
Friend leukemia integration 1 |
| chr15_-_100322089 | 1.60 |
ENSMUST00000154331.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr7_-_119078472 | 1.58 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr10_+_79656823 | 1.57 |
ENSMUST00000169041.9
|
Misp
|
mitotic spindle positioning |
| chr19_+_11495858 | 1.54 |
ENSMUST00000025580.10
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr4_-_47474283 | 1.52 |
ENSMUST00000044148.3
|
Alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr2_-_93292734 | 1.51 |
ENSMUST00000099696.8
|
Cd82
|
CD82 antigen |
| chr5_+_145104011 | 1.51 |
ENSMUST00000160629.8
ENSMUST00000070487.12 ENSMUST00000160422.8 ENSMUST00000162244.8 |
Cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr11_+_101137231 | 1.50 |
ENSMUST00000122006.8
ENSMUST00000151830.2 |
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr11_+_115824108 | 1.49 |
ENSMUST00000140991.2
|
Sap30bp
|
SAP30 binding protein |
| chr19_-_5660057 | 1.49 |
ENSMUST00000236229.2
ENSMUST00000235701.2 ENSMUST00000236264.2 |
Kat5
|
K(lysine) acetyltransferase 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 32.6 | GO:0030221 | basophil differentiation(GO:0030221) |
| 4.2 | 25.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 3.7 | 18.6 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 3.7 | 25.8 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 3.0 | 15.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 2.1 | 6.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 2.0 | 9.8 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 1.9 | 5.6 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 1.8 | 66.0 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 1.7 | 6.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.6 | 4.8 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 1.6 | 4.8 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.5 | 41.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 1.4 | 4.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 1.4 | 4.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 1.2 | 3.6 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.2 | 3.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 1.2 | 5.8 | GO:2001274 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 1.1 | 6.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.1 | 3.4 | GO:0072248 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.1 | 7.8 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 1.0 | 3.9 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 1.0 | 6.7 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 1.0 | 2.9 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 1.0 | 6.7 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.9 | 5.6 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.9 | 6.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.9 | 8.7 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.9 | 2.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.8 | 3.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.8 | 4.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.7 | 0.7 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.7 | 3.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.7 | 5.8 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.7 | 6.5 | GO:0061092 | involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.7 | 2.1 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.7 | 6.7 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.7 | 5.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.6 | 4.9 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.6 | 2.5 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.6 | 2.3 | GO:1903944 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.6 | 10.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.6 | 10.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.6 | 1.7 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.5 | 15.8 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.5 | 2.7 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.5 | 2.2 | GO:0072021 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.5 | 3.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.5 | 4.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 2.0 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.5 | 1.5 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.5 | 9.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.5 | 1.5 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.5 | 4.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 | 1.7 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.4 | 2.5 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.4 | 12.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 2.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 2.3 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.4 | 2.9 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.4 | 6.8 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.4 | 2.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.3 | 1.0 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.3 | 2.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 9.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.3 | 2.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.3 | 2.3 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.3 | 1.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.3 | 1.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 1.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 0.8 | GO:0072422 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.3 | 1.3 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.2 | 2.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.2 | 2.6 | GO:1901249 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.2 | 31.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 10.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.2 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 1.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 12.9 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 4.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 2.9 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.2 | 3.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 1.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 19.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 1.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 2.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.2 | 10.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 0.3 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.2 | 0.5 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.7 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.3 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.1 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 2.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 2.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 3.6 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 4.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.5 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 1.7 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.7 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 1.3 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.1 | 2.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 1.0 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 11.2 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.1 | 3.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 8.6 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 1.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.1 | 3.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 2.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 3.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 2.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 3.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 12.0 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 4.4 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.1 | 4.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 6.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.5 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 3.5 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.0 | 3.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 3.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.2 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 1.5 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 3.0 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 2.1 | GO:0032272 | negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 3.9 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 25.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 2.3 | 6.8 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 2.1 | 12.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.4 | 13.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.3 | 7.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.2 | 4.8 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 1.1 | 15.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.0 | 4.9 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.0 | 18.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.9 | 3.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.8 | 11.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.6 | 9.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.6 | 3.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.5 | 5.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.5 | 2.9 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.5 | 2.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.5 | 12.8 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.4 | 1.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 2.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.4 | 5.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 9.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 11.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 41.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 3.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 5.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 31.3 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 2.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 9.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 1.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 31.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 7.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 2.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 2.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 78.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 5.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 3.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.9 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 4.1 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 20.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 2.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 2.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 6.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 10.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 5.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 15.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 5.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.6 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.1 | 1.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 5.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 8.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 9.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 3.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 1.9 | 41.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 1.8 | 32.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 1.6 | 15.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.4 | 8.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 1.4 | 15.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.4 | 5.6 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 1.3 | 11.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 1.3 | 8.9 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.2 | 6.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 1.2 | 4.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.1 | 12.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.1 | 7.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.0 | 7.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.0 | 3.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 1.0 | 4.8 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.9 | 11.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.9 | 2.7 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.9 | 4.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.8 | 2.5 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.7 | 6.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.7 | 2.9 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.7 | 5.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.7 | 9.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 9.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.6 | 3.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 2.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.6 | 4.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 1.8 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.6 | 11.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.6 | 1.7 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.5 | 4.7 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.5 | 2.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.5 | 1.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.5 | 7.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.5 | 2.3 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.4 | 18.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 6.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 3.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 2.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 1.7 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 1.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 6.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 25.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 8.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.3 | 20.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 3.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 2.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 8.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 31.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 6.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 2.8 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 19.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 8.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 2.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 4.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 2.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 3.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 0.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 3.0 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 12.0 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 7.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 16.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 2.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 2.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 13.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 4.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 6.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 8.9 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 1.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 4.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 36.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 4.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 2.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 2.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 5.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 18.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.9 | 68.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 5.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.4 | 34.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 8.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 23.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 9.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 3.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 5.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 6.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 6.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 11.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 7.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 6.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 11.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 1.4 | 62.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.7 | 18.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 8.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 26.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 9.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 25.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 18.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.3 | 50.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 12.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.3 | 3.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 38.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 8.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 13.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 6.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 6.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 4.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 12.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 7.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 4.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 4.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.7 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 3.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 5.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 4.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |