Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
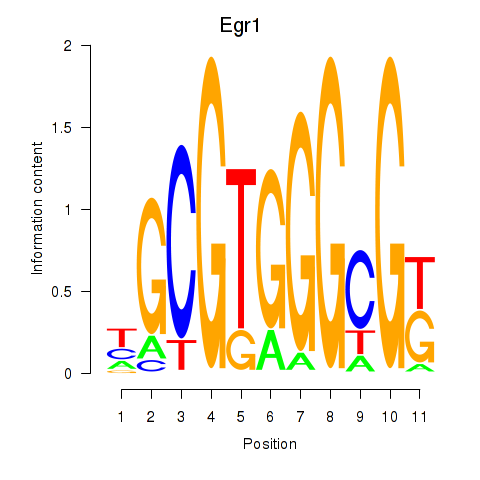
Results for Egr1
Z-value: 1.97
Transcription factors associated with Egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr1
|
ENSMUSG00000038418.8 | Egr1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr1 | mm39_v1_chr18_+_34994253_34994268 | -0.07 | 6.9e-01 | Click! |
Activity profile of Egr1 motif
Sorted Z-values of Egr1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_109419371 | 11.61 |
ENSMUST00000109844.11
ENSMUST00000109842.9 ENSMUST00000109843.8 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr12_-_76756772 | 9.99 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr12_+_109419575 | 9.53 |
ENSMUST00000173539.8
ENSMUST00000109841.9 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr10_-_80413119 | 7.45 |
ENSMUST00000038558.9
|
Klf16
|
Kruppel-like factor 16 |
| chr12_+_109419454 | 7.10 |
ENSMUST00000109846.11
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr6_-_60805873 | 6.98 |
ENSMUST00000114268.5
|
Snca
|
synuclein, alpha |
| chr6_+_121613177 | 6.96 |
ENSMUST00000032203.9
|
A2m
|
alpha-2-macroglobulin |
| chr14_-_70873385 | 6.80 |
ENSMUST00000228295.2
ENSMUST00000022695.16 |
Dmtn
|
dematin actin binding protein |
| chr3_-_88417251 | 6.45 |
ENSMUST00000149068.2
|
Lmna
|
lamin A |
| chr16_+_93680783 | 6.39 |
ENSMUST00000023666.11
ENSMUST00000117099.8 |
Chaf1b
|
chromatin assembly factor 1, subunit B (p60) |
| chr7_-_142213219 | 6.37 |
ENSMUST00000121128.8
|
Igf2
|
insulin-like growth factor 2 |
| chr2_+_117942357 | 5.92 |
ENSMUST00000039559.9
|
Thbs1
|
thrombospondin 1 |
| chr12_-_32111214 | 5.80 |
ENSMUST00000003079.12
ENSMUST00000036497.16 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chr17_+_37180437 | 5.75 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr9_-_37464200 | 5.43 |
ENSMUST00000065668.12
|
Nrgn
|
neurogranin |
| chr3_-_100396635 | 5.39 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr7_-_126303947 | 5.32 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr3_-_127689890 | 4.76 |
ENSMUST00000057198.9
ENSMUST00000199273.2 |
Fam241a
|
family with sequence similarity 241, member A |
| chrX_+_7708295 | 4.71 |
ENSMUST00000115667.10
ENSMUST00000115668.10 ENSMUST00000115665.2 |
Otud5
|
OTU domain containing 5 |
| chr4_-_43045685 | 4.59 |
ENSMUST00000107956.8
ENSMUST00000107957.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr4_+_118965908 | 4.53 |
ENSMUST00000030398.10
|
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr10_-_81214293 | 4.50 |
ENSMUST00000140901.8
|
Fzr1
|
fizzy and cell division cycle 20 related 1 |
| chr7_-_126303351 | 4.39 |
ENSMUST00000106364.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr4_-_43046196 | 4.20 |
ENSMUST00000036462.12
|
Fam214b
|
family with sequence similarity 214, member B |
| chr4_+_118965992 | 3.94 |
ENSMUST00000134105.8
ENSMUST00000144329.8 ENSMUST00000208090.2 |
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr7_-_126014027 | 3.82 |
ENSMUST00000032968.7
ENSMUST00000206325.2 |
Cd19
|
CD19 antigen |
| chr6_+_142702403 | 3.74 |
ENSMUST00000032419.9
|
Cmas
|
cytidine monophospho-N-acetylneuraminic acid synthetase |
| chr8_+_84724130 | 3.70 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr7_-_37806912 | 3.50 |
ENSMUST00000108023.10
|
Ccne1
|
cyclin E1 |
| chr16_+_49676130 | 3.39 |
ENSMUST00000230641.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr10_-_12839995 | 3.36 |
ENSMUST00000219727.2
ENSMUST00000163425.9 ENSMUST00000042861.7 ENSMUST00000218685.2 |
Stx11
|
syntaxin 11 |
| chr13_-_55661240 | 3.31 |
ENSMUST00000069929.13
ENSMUST00000069968.13 ENSMUST00000131306.8 ENSMUST00000046246.13 |
Pdlim7
|
PDZ and LIM domain 7 |
| chrX_-_74174608 | 3.28 |
ENSMUST00000033775.9
|
Mpp1
|
membrane protein, palmitoylated |
| chrX_-_74174450 | 3.18 |
ENSMUST00000114092.8
ENSMUST00000132501.8 ENSMUST00000153318.8 ENSMUST00000155742.2 |
Mpp1
|
membrane protein, palmitoylated |
| chr1_+_135060431 | 3.07 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr9_+_62746055 | 3.06 |
ENSMUST00000034776.13
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr3_-_86827640 | 3.04 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr7_+_120442773 | 3.00 |
ENSMUST00000143279.3
ENSMUST00000106489.9 |
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr7_-_126625617 | 2.93 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr7_+_99184645 | 2.92 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr4_-_133615075 | 2.90 |
ENSMUST00000003741.16
ENSMUST00000105894.11 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chrX_+_7708034 | 2.90 |
ENSMUST00000033494.16
ENSMUST00000115666.8 |
Otud5
|
OTU domain containing 5 |
| chr1_-_59012579 | 2.86 |
ENSMUST00000173590.2
ENSMUST00000027186.12 |
Trak2
|
trafficking protein, kinesin binding 2 |
| chr15_+_79231720 | 2.85 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr5_-_33432310 | 2.80 |
ENSMUST00000201372.3
ENSMUST00000202962.4 ENSMUST00000201575.4 ENSMUST00000202868.4 ENSMUST00000079746.10 |
Ctbp1
|
C-terminal binding protein 1 |
| chr2_+_118644717 | 2.74 |
ENSMUST00000028803.14
ENSMUST00000126045.8 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr16_-_18448614 | 2.72 |
ENSMUST00000231956.2
ENSMUST00000096987.7 |
Septin5
|
septin 5 |
| chrX_+_161543384 | 2.69 |
ENSMUST00000033720.12
ENSMUST00000112327.8 |
Rbbp7
|
retinoblastoma binding protein 7, chromatin remodeling factor |
| chrX_+_9138995 | 2.68 |
ENSMUST00000015486.7
|
Xk
|
X-linked Kx blood group |
| chr16_+_49675969 | 2.68 |
ENSMUST00000229101.2
ENSMUST00000230836.2 ENSMUST00000229206.2 ENSMUST00000084838.14 ENSMUST00000230281.2 |
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr11_+_117006020 | 2.57 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chrX_-_74174524 | 2.55 |
ENSMUST00000114091.8
|
Mpp1
|
membrane protein, palmitoylated |
| chr15_-_36609208 | 2.55 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr10_-_80269436 | 2.54 |
ENSMUST00000105346.10
ENSMUST00000020377.13 ENSMUST00000105340.8 ENSMUST00000020379.13 ENSMUST00000105344.8 ENSMUST00000105342.8 ENSMUST00000105345.10 ENSMUST00000105343.8 |
Tcf3
|
transcription factor 3 |
| chr9_-_57169830 | 2.52 |
ENSMUST00000215298.2
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr9_-_57169872 | 2.50 |
ENSMUST00000213199.2
ENSMUST00000034846.7 |
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr15_-_64184485 | 2.49 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr6_-_70769135 | 2.49 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr9_+_44245981 | 2.45 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr9_+_106080307 | 2.42 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr4_-_133856025 | 2.42 |
ENSMUST00000105879.2
ENSMUST00000030651.9 |
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3 |
| chr2_+_118644675 | 2.37 |
ENSMUST00000110842.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr4_-_133694607 | 2.35 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr15_+_88746380 | 2.34 |
ENSMUST00000042818.11
|
Pim3
|
proviral integration site 3 |
| chr14_-_47514248 | 2.32 |
ENSMUST00000187531.8
ENSMUST00000111790.2 |
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr7_+_26958150 | 2.32 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr12_+_85645801 | 2.30 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chrX_+_161543423 | 2.28 |
ENSMUST00000112326.8
|
Rbbp7
|
retinoblastoma binding protein 7, chromatin remodeling factor |
| chr11_+_117005958 | 2.24 |
ENSMUST00000021177.15
|
Sec14l1
|
SEC14-like lipid binding 1 |
| chr8_-_123278054 | 2.20 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr12_+_85646162 | 2.20 |
ENSMUST00000050687.14
|
Jdp2
|
Jun dimerization protein 2 |
| chr12_+_108602008 | 2.18 |
ENSMUST00000172409.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr7_-_126625657 | 2.17 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chrX_-_72703330 | 2.08 |
ENSMUST00000114473.8
ENSMUST00000002087.14 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr13_+_55593116 | 2.08 |
ENSMUST00000001115.16
ENSMUST00000224995.2 ENSMUST00000225925.2 ENSMUST00000099482.5 ENSMUST00000224118.2 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr6_+_72074545 | 2.07 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr12_-_36206780 | 2.02 |
ENSMUST00000223382.2
ENSMUST00000020856.6 |
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr5_-_110987604 | 2.01 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr12_+_108601963 | 1.99 |
ENSMUST00000223109.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr2_+_118644475 | 1.99 |
ENSMUST00000134661.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr5_+_143608194 | 1.98 |
ENSMUST00000116456.10
|
Cyth3
|
cytohesin 3 |
| chr4_-_133694543 | 1.97 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr15_+_81119700 | 1.94 |
ENSMUST00000166855.3
|
Mchr1
|
melanin-concentrating hormone receptor 1 |
| chr6_+_72074718 | 1.93 |
ENSMUST00000187007.3
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr11_-_120238917 | 1.92 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr14_+_24540745 | 1.91 |
ENSMUST00000112384.10
|
Rps24
|
ribosomal protein S24 |
| chr5_-_107873883 | 1.90 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr7_+_29991101 | 1.90 |
ENSMUST00000150892.2
ENSMUST00000126216.2 ENSMUST00000014065.16 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr15_+_76131020 | 1.90 |
ENSMUST00000229380.2
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr9_+_54606832 | 1.86 |
ENSMUST00000070070.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr2_-_34262012 | 1.85 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr14_+_24540777 | 1.84 |
ENSMUST00000169826.3
ENSMUST00000225023.2 ENSMUST00000223999.2 |
Rps24
|
ribosomal protein S24 |
| chr5_-_137530214 | 1.84 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr18_-_77855446 | 1.80 |
ENSMUST00000048192.9
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr5_-_148988413 | 1.78 |
ENSMUST00000093196.11
|
Hmgb1
|
high mobility group box 1 |
| chr5_-_114046746 | 1.78 |
ENSMUST00000004646.13
|
Coro1c
|
coronin, actin binding protein 1C |
| chr4_-_93223746 | 1.77 |
ENSMUST00000066774.6
|
Tusc1
|
tumor suppressor candidate 1 |
| chr7_-_16007542 | 1.75 |
ENSMUST00000169612.3
|
Inafm1
|
InaF motif containing 1 |
| chr7_+_127566629 | 1.74 |
ENSMUST00000106251.10
ENSMUST00000077609.12 ENSMUST00000121616.9 |
Fus
|
fused in sarcoma |
| chr8_+_85753452 | 1.74 |
ENSMUST00000047281.10
|
Trir
|
telomerase RNA component interacting RNase |
| chr7_+_139673300 | 1.69 |
ENSMUST00000026540.9
|
Prap1
|
proline-rich acidic protein 1 |
| chr3_-_83947416 | 1.68 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr7_+_27186335 | 1.67 |
ENSMUST00000008528.8
|
Sertad1
|
SERTA domain containing 1 |
| chr15_+_78784043 | 1.65 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr13_+_55517545 | 1.65 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr3_+_103009920 | 1.64 |
ENSMUST00000173206.8
|
Dennd2c
|
DENN/MADD domain containing 2C |
| chr15_+_78783867 | 1.64 |
ENSMUST00000134703.8
ENSMUST00000061239.14 ENSMUST00000109698.9 |
Gm49510
Sh3bp1
|
predicted gene, 49510 SH3-domain binding protein 1 |
| chr1_-_180641430 | 1.63 |
ENSMUST00000162814.8
|
H3f3a
|
H3.3 histone A |
| chr14_+_24540815 | 1.63 |
ENSMUST00000224568.2
|
Rps24
|
ribosomal protein S24 |
| chr4_-_135714465 | 1.62 |
ENSMUST00000105851.9
|
Pithd1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr4_+_131600918 | 1.57 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr9_+_54606144 | 1.53 |
ENSMUST00000120452.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr6_+_28423539 | 1.52 |
ENSMUST00000020717.12
ENSMUST00000169841.2 |
Arf5
|
ADP-ribosylation factor 5 |
| chr7_-_126625739 | 1.51 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr10_-_80156337 | 1.51 |
ENSMUST00000020341.9
|
2310011J03Rik
|
RIKEN cDNA 2310011J03 gene |
| chrX_-_72703652 | 1.50 |
ENSMUST00000114472.8
|
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr14_+_51162635 | 1.50 |
ENSMUST00000128395.2
|
Apex1
|
apurinic/apyrimidinic endonuclease 1 |
| chrX_+_48559327 | 1.48 |
ENSMUST00000114904.10
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr1_-_75119277 | 1.47 |
ENSMUST00000168720.8
ENSMUST00000041213.12 ENSMUST00000189809.2 |
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr9_+_54606798 | 1.46 |
ENSMUST00000154690.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr5_-_110987441 | 1.45 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr6_+_4747298 | 1.45 |
ENSMUST00000166678.2
ENSMUST00000176204.8 |
Peg10
|
paternally expressed 10 |
| chr16_-_18448454 | 1.45 |
ENSMUST00000231622.2
|
Septin5
|
septin 5 |
| chr17_+_29020064 | 1.44 |
ENSMUST00000004985.11
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr12_-_36206750 | 1.44 |
ENSMUST00000221388.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr19_+_6952580 | 1.44 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr11_-_100331754 | 1.43 |
ENSMUST00000107397.2
ENSMUST00000092689.9 |
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr1_-_97589365 | 1.42 |
ENSMUST00000153115.8
ENSMUST00000142234.2 |
Macir
|
macrophage immunometabolism regulator |
| chr2_+_69652714 | 1.41 |
ENSMUST00000053087.4
|
Klhl23
|
kelch-like 23 |
| chr4_-_133695204 | 1.39 |
ENSMUST00000100472.10
ENSMUST00000136327.2 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr8_+_40876827 | 1.37 |
ENSMUST00000049389.11
ENSMUST00000128166.8 ENSMUST00000167766.2 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr5_+_100187844 | 1.37 |
ENSMUST00000169390.8
ENSMUST00000031268.8 |
Enoph1
|
enolase-phosphatase 1 |
| chr2_-_34803988 | 1.37 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr2_+_145009625 | 1.36 |
ENSMUST00000110007.8
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr7_-_126102579 | 1.35 |
ENSMUST00000040202.15
|
Atxn2l
|
ataxin 2-like |
| chr4_+_47474652 | 1.32 |
ENSMUST00000065678.6
|
Sec61b
|
Sec61 beta subunit |
| chr17_+_35113490 | 1.32 |
ENSMUST00000052778.10
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr19_+_5618029 | 1.31 |
ENSMUST00000235575.2
ENSMUST00000235542.2 |
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr1_-_181039509 | 1.30 |
ENSMUST00000162819.9
ENSMUST00000237749.2 |
Wdr26
|
WD repeat domain 26 |
| chr6_-_88818832 | 1.30 |
ENSMUST00000032169.8
|
Abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr16_+_93680846 | 1.30 |
ENSMUST00000142316.2
|
Chaf1b
|
chromatin assembly factor 1, subunit B (p60) |
| chrX_-_156275231 | 1.29 |
ENSMUST00000112529.8
|
Sms
|
spermine synthase |
| chr6_-_30936013 | 1.29 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr19_+_6952319 | 1.28 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr11_+_70323452 | 1.27 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr15_+_76130947 | 1.27 |
ENSMUST00000229772.2
ENSMUST00000230347.2 ENSMUST00000023225.8 |
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr13_-_98453475 | 1.25 |
ENSMUST00000022163.15
ENSMUST00000152704.8 |
Btf3
|
basic transcription factor 3 |
| chr6_+_29433247 | 1.25 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr7_-_121620366 | 1.23 |
ENSMUST00000033160.15
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr15_-_76594723 | 1.23 |
ENSMUST00000036852.9
|
Recql4
|
RecQ protein-like 4 |
| chr6_-_4747157 | 1.22 |
ENSMUST00000126151.8
ENSMUST00000115577.9 ENSMUST00000101677.9 ENSMUST00000115579.8 ENSMUST00000004750.15 |
Sgce
|
sarcoglycan, epsilon |
| chr5_-_108515740 | 1.22 |
ENSMUST00000197216.3
|
Gm42517
|
predicted gene 42517 |
| chr8_+_34222058 | 1.21 |
ENSMUST00000167264.8
ENSMUST00000187392.7 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr9_+_108673171 | 1.20 |
ENSMUST00000195514.6
ENSMUST00000085018.6 ENSMUST00000192028.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr6_+_85408953 | 1.17 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr4_-_133695264 | 1.17 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr18_+_82932747 | 1.15 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr8_+_34221861 | 1.15 |
ENSMUST00000170705.8
|
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr14_-_121616616 | 1.14 |
ENSMUST00000079817.8
|
Stk24
|
serine/threonine kinase 24 |
| chr4_-_152216322 | 1.14 |
ENSMUST00000105653.8
|
Espn
|
espin |
| chr11_-_116001037 | 1.13 |
ENSMUST00000106441.8
ENSMUST00000021120.6 |
Trim47
|
tripartite motif-containing 47 |
| chr10_-_45346297 | 1.12 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr7_-_126102470 | 1.11 |
ENSMUST00000206577.2
|
Atxn2l
|
ataxin 2-like |
| chr5_-_34093678 | 1.11 |
ENSMUST00000030993.8
|
Nelfa
|
negative elongation factor complex member A, Whsc2 |
| chr13_+_21938258 | 1.11 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr2_+_25070749 | 1.10 |
ENSMUST00000104999.4
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr5_-_137609634 | 1.10 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr2_+_180231038 | 1.10 |
ENSMUST00000029087.4
|
Ogfr
|
opioid growth factor receptor |
| chr8_+_34222266 | 1.08 |
ENSMUST00000190675.2
ENSMUST00000171010.8 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chrX_+_48559432 | 1.05 |
ENSMUST00000042444.7
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr19_+_5618096 | 1.04 |
ENSMUST00000096318.4
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr1_+_86514822 | 1.03 |
ENSMUST00000027446.11
|
Cops7b
|
COP9 signalosome subunit 7B |
| chr17_-_31877703 | 1.03 |
ENSMUST00000236475.2
ENSMUST00000166526.9 ENSMUST00000014684.6 |
U2af1
|
U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 1 |
| chr6_+_85428464 | 1.02 |
ENSMUST00000032078.9
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta) |
| chr1_+_75358758 | 1.02 |
ENSMUST00000148515.8
ENSMUST00000113590.8 |
Speg
|
SPEG complex locus |
| chr12_-_69629758 | 1.02 |
ENSMUST00000058639.11
|
Vcpkmt
|
valosin containing protein lysine (K) methyltransferase |
| chr14_+_33645539 | 1.02 |
ENSMUST00000168727.3
|
Gdf10
|
growth differentiation factor 10 |
| chr15_-_76594625 | 1.01 |
ENSMUST00000230544.2
|
Recql4
|
RecQ protein-like 4 |
| chr7_-_51655426 | 1.01 |
ENSMUST00000209193.2
|
Svip
|
small VCP/p97-interacting protein |
| chr8_-_81466126 | 1.01 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr2_-_181101158 | 1.00 |
ENSMUST00000155535.2
ENSMUST00000029106.13 ENSMUST00000087409.10 |
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr1_-_74990821 | 1.00 |
ENSMUST00000164097.4
|
Ihh
|
Indian hedgehog |
| chr8_+_110595216 | 0.99 |
ENSMUST00000179721.8
ENSMUST00000034175.5 |
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr1_-_96799832 | 0.99 |
ENSMUST00000071985.6
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr11_-_96668318 | 0.97 |
ENSMUST00000127375.2
ENSMUST00000021246.9 ENSMUST00000107661.10 |
Snx11
|
sorting nexin 11 |
| chr5_-_137609691 | 0.97 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr9_-_119812042 | 0.97 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr1_-_52272370 | 0.97 |
ENSMUST00000114513.9
ENSMUST00000114510.8 |
Gls
|
glutaminase |
| chr9_-_44145309 | 0.97 |
ENSMUST00000206720.2
|
Cbl
|
Casitas B-lineage lymphoma |
| chrX_+_55777139 | 0.96 |
ENSMUST00000023854.10
ENSMUST00000114769.9 |
Fhl1
|
four and a half LIM domains 1 |
| chr2_+_78699360 | 0.96 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr14_+_54713557 | 0.95 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr11_-_4696778 | 0.95 |
ENSMUST00000009219.3
|
Cabp7
|
calcium binding protein 7 |
| chr17_+_24689366 | 0.95 |
ENSMUST00000053024.8
|
Pgp
|
phosphoglycolate phosphatase |
| chr9_-_90152759 | 0.95 |
ENSMUST00000041767.14
ENSMUST00000128874.3 |
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr11_-_6015538 | 0.93 |
ENSMUST00000101585.10
ENSMUST00000066431.14 ENSMUST00000109815.9 ENSMUST00000109812.9 ENSMUST00000101586.3 ENSMUST00000093355.12 ENSMUST00000019133.11 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr15_-_36283244 | 0.93 |
ENSMUST00000228358.2
ENSMUST00000022890.10 |
Rnf19a
|
ring finger protein 19A |
| chr5_+_33978035 | 0.93 |
ENSMUST00000075812.11
ENSMUST00000114397.9 ENSMUST00000155880.8 |
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr5_+_145020640 | 0.92 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr3_+_84859453 | 0.90 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 2.3 | 7.0 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 2.0 | 5.9 | GO:0010752 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of cGMP-mediated signaling(GO:0010752) |
| 1.9 | 7.6 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 1.7 | 8.5 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 1.2 | 5.0 | GO:0010286 | heat acclimation(GO:0010286) |
| 1.2 | 9.7 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 1.1 | 6.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.1 | 6.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 2.8 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.9 | 3.7 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.7 | 26.9 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.7 | 7.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.7 | 4.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.6 | 2.5 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.6 | 7.3 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.6 | 6.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.6 | 1.8 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.6 | 6.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.5 | 4.4 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.5 | 1.9 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.5 | 2.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.5 | 5.0 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.4 | 2.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 3.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.4 | 1.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.4 | 2.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.4 | 2.5 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.4 | 2.5 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.4 | 2.7 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.4 | 1.1 | GO:0072708 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.4 | 2.6 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 2.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 1.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 0.3 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.3 | 7.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 1.0 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.3 | 1.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.6 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.3 | 2.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 6.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 2.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.3 | 0.9 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.3 | 3.4 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.3 | 4.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 0.9 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.3 | 1.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.3 | 4.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 2.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 2.5 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.2 | 10.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 3.5 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 2.4 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.2 | 5.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.2 | 7.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 0.7 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.2 | 0.9 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.2 | 3.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 9.0 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.2 | 1.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.6 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 0.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 1.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.4 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 2.3 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.2 | 0.5 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.2 | 0.5 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.2 | 1.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.2 | 0.5 | GO:1990869 | beta selection(GO:0043366) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.2 | 1.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 1.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.6 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 2.7 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.4 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.1 | 4.9 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 1.8 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 2.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.6 | GO:0036302 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) atrioventricular canal development(GO:0036302) mammary placode formation(GO:0060596) |
| 0.1 | 5.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 3.0 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 1.5 | GO:0043247 | telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.2 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 7.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.9 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 1.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 3.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 1.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 1.8 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 1.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 1.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 0.8 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.5 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.6 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.1 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.8 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.6 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.0 | GO:1904867 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 4.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 4.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.8 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.7 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.1 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 1.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.8 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.5 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.9 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.6 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.9 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.3 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 1.9 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0032962 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.9 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 4.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.0 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 0.3 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.9 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 2.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 2.6 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 1.1 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.5 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.4 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.6 | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity(GO:1901020) |
| 0.0 | 1.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) regulation of neurotrophin production(GO:0032899) positive regulation of neurotrophin production(GO:0032901) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 1.7 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 1.0 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 1.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.7 | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 1.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.5 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.6 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.4 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.1 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 1.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.8 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 1.0 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.1 | GO:1900113 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.7 | 7.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.2 | 10.0 | GO:0008091 | spectrin(GO:0008091) |
| 1.1 | 3.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 1.0 | 6.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.9 | 6.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.7 | 5.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 1.6 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.5 | 8.5 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.5 | 7.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 10.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 1.6 | GO:0001740 | Barr body(GO:0001740) |
| 0.4 | 6.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 1.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.3 | 2.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 1.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 1.8 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 0.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 1.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) endoplasmic reticulum Sec complex(GO:0031205) translocon complex(GO:0071256) |
| 0.2 | 5.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 1.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 0.5 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 1.0 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 6.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.1 | 4.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 2.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 5.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 3.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.6 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 7.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 3.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 3.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 21.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.7 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 3.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.8 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.2 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 3.3 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 1.7 | 8.5 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 1.5 | 5.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.3 | 4.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.0 | 4.2 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 1.0 | 3.0 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.8 | 6.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.8 | 4.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.6 | 7.0 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.6 | 1.8 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.6 | 6.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.5 | 1.5 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.5 | 7.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 9.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 2.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.4 | 3.7 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.4 | 2.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.4 | 2.2 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 0.3 | 1.6 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 1.0 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.3 | 2.8 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 2.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 10.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 0.7 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 1.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 2.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 1.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 0.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.2 | 5.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 0.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 7.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 6.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 0.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 1.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 3.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 1.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.4 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 3.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 2.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 3.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.6 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.9 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.8 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) mRNA CDS binding(GO:1990715) |
| 0.1 | 5.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.5 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 2.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 7.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 2.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 11.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 1.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 2.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 9.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.1 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 2.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 13.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 3.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.0 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 25.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 9.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 13.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 3.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 3.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 9.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 6.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 6.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 5.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 5.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 31.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.5 | 8.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 7.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 6.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 6.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 6.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 6.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 3.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 1.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 8.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 4.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 7.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 6.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 4.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 6.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 7.9 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.1 | 3.9 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.1 | 1.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 5.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 3.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 3.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 3.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 5.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 3.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |