Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
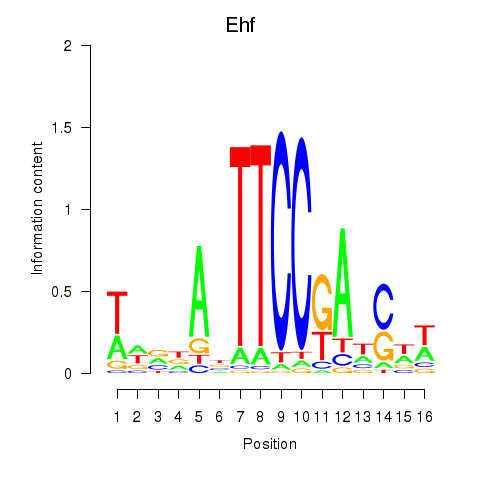
Results for Ehf
Z-value: 0.39
Transcription factors associated with Ehf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ehf
|
ENSMUSG00000012350.16 | Ehf |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ehf | mm39_v1_chr2_-_103114105_103114147 | -0.32 | 6.0e-02 | Click! |
Activity profile of Ehf motif
Sorted Z-values of Ehf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ehf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_83421333 | 1.32 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr12_-_104831266 | 1.19 |
ENSMUST00000109937.9
|
Clmn
|
calmin |
| chr6_+_54249817 | 0.99 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr11_-_95966407 | 0.87 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr9_+_106324952 | 0.83 |
ENSMUST00000215475.2
ENSMUST00000187106.7 ENSMUST00000190167.7 |
Abhd14b
|
abhydrolase domain containing 14b |
| chr11_-_95966477 | 0.74 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr9_+_107419416 | 0.73 |
ENSMUST00000010201.9
|
Nprl2
|
NPR2 like, GATOR1 complex subunit |
| chr11_+_69286473 | 0.63 |
ENSMUST00000144531.2
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr11_-_69553390 | 0.52 |
ENSMUST00000129224.8
ENSMUST00000155200.8 |
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr11_-_69553451 | 0.51 |
ENSMUST00000018905.12
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr9_-_107419309 | 0.32 |
ENSMUST00000195235.6
|
Cyb561d2
|
cytochrome b-561 domain containing 2 |
| chr13_-_91372072 | 0.24 |
ENSMUST00000022119.6
|
Atg10
|
autophagy related 10 |
| chr11_-_54751738 | 0.23 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chr19_+_6214416 | 0.19 |
ENSMUST00000045042.8
ENSMUST00000237511.2 |
Batf2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr9_-_107419015 | 0.19 |
ENSMUST00000041459.9
|
Cyb561d2
|
cytochrome b-561 domain containing 2 |
| chr16_+_16120810 | 0.17 |
ENSMUST00000159962.8
ENSMUST00000059955.15 |
Yars2
|
tyrosyl-tRNA synthetase 2 (mitochondrial) |
| chrX_+_152341587 | 0.15 |
ENSMUST00000112573.8
ENSMUST00000056754.4 |
Cypt3
|
cysteine-rich perinuclear theca 3 |
| chr11_-_121095462 | 0.14 |
ENSMUST00000026169.7
|
Ogfod3
|
2-oxoglutarate and iron-dependent oxygenase domain containing 3 |
| chr15_+_4404965 | 0.14 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr11_-_69286159 | 0.13 |
ENSMUST00000108660.8
ENSMUST00000051620.5 |
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr18_+_24737009 | 0.12 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr4_-_120672900 | 0.12 |
ENSMUST00000120779.8
|
Nfyc
|
nuclear transcription factor-Y gamma |
| chr3_-_116388334 | 0.12 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr6_-_130170075 | 0.10 |
ENSMUST00000112032.8
ENSMUST00000071554.3 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr9_+_32027335 | 0.10 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chrX_+_70707271 | 0.09 |
ENSMUST00000070449.6
|
Gpr50
|
G-protein-coupled receptor 50 |
| chr6_-_30892508 | 0.08 |
ENSMUST00000048580.2
|
Tsga13
|
testis specific gene A13 |
| chr16_-_19105587 | 0.07 |
ENSMUST00000056727.5
|
Olfr164
|
olfactory receptor 164 |
| chr1_+_127796508 | 0.04 |
ENSMUST00000037649.6
ENSMUST00000212506.2 |
Rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr2_-_164646794 | 0.04 |
ENSMUST00000103094.11
ENSMUST00000017451.7 |
Acot8
|
acyl-CoA thioesterase 8 |
| chr18_+_37610858 | 0.04 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr9_-_106324642 | 0.03 |
ENSMUST00000185334.7
ENSMUST00000187001.2 ENSMUST00000171678.9 ENSMUST00000190798.7 ENSMUST00000048685.13 ENSMUST00000171925.8 |
Abhd14a
|
abhydrolase domain containing 14A |
| chr13_-_8921000 | 0.02 |
ENSMUST00000164183.9
|
Wdr37
|
WD repeat domain 37 |
| chr1_+_174196485 | 0.02 |
ENSMUST00000085862.2
|
Olfr417
|
olfactory receptor 417 |
| chr9_+_77543776 | 0.01 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:1903233 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |