Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
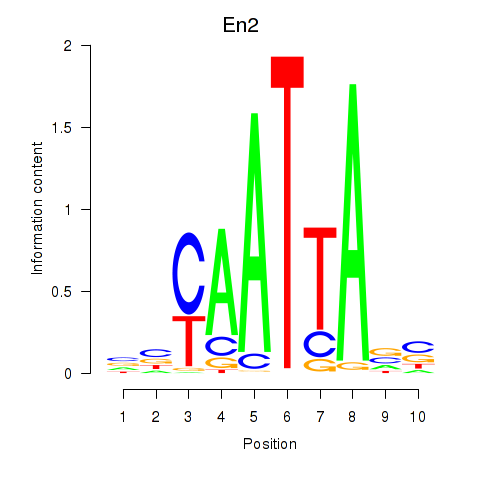
Results for En2
Z-value: 1.12
Transcription factors associated with En2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En2
|
ENSMUSG00000039095.9 | En2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En2 | mm39_v1_chr5_+_28370687_28370720 | 0.15 | 3.8e-01 | Click! |
Activity profile of En2 motif
Sorted Z-values of En2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of En2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_127734384 | 4.79 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_+_127657142 | 3.61 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr3_+_59989282 | 2.94 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr14_+_55797468 | 2.82 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr17_-_84154196 | 2.59 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr17_-_84154173 | 2.59 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr14_+_55797934 | 2.52 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr7_+_51528788 | 2.40 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr12_-_84497718 | 2.27 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr19_-_7943365 | 2.25 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr14_+_55798362 | 2.01 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr5_-_77262968 | 2.00 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr5_+_135135735 | 1.81 |
ENSMUST00000201977.4
ENSMUST00000005507.10 |
Mlxipl
|
MLX interacting protein-like |
| chrM_+_10167 | 1.74 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr2_+_30254239 | 1.63 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chr14_+_55797443 | 1.47 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr2_+_118692435 | 1.44 |
ENSMUST00000028807.6
|
Ivd
|
isovaleryl coenzyme A dehydrogenase |
| chr10_-_126956991 | 1.41 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr12_-_108145498 | 1.38 |
ENSMUST00000071095.14
|
Setd3
|
SET domain containing 3 |
| chr6_-_54969843 | 1.33 |
ENSMUST00000203208.2
|
Ggct
|
gamma-glutamyl cyclotransferase |
| chrM_+_9870 | 1.16 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr3_-_54962899 | 1.15 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr6_-_54969928 | 1.15 |
ENSMUST00000131475.2
|
Ggct
|
gamma-glutamyl cyclotransferase |
| chrM_+_3906 | 1.10 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr3_+_63203516 | 1.06 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chrM_+_11735 | 1.04 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr18_+_84869456 | 1.03 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr8_-_45835234 | 1.01 |
ENSMUST00000093526.13
|
Fam149a
|
family with sequence similarity 149, member A |
| chr15_+_100251030 | 0.99 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr13_+_23728222 | 0.96 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr7_-_119494669 | 0.93 |
ENSMUST00000098080.9
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr12_+_111780604 | 0.93 |
ENSMUST00000021714.9
ENSMUST00000223211.2 ENSMUST00000222843.2 ENSMUST00000221375.2 |
Zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr7_+_44240310 | 0.92 |
ENSMUST00000107906.6
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr11_-_115078147 | 0.91 |
ENSMUST00000103038.8
ENSMUST00000103039.2 ENSMUST00000103040.11 |
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr16_+_22676589 | 0.89 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr2_-_147887810 | 0.88 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chr19_-_42117420 | 0.88 |
ENSMUST00000161873.2
ENSMUST00000018965.4 |
Avpi1
|
arginine vasopressin-induced 1 |
| chrM_+_9459 | 0.85 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_-_86884507 | 0.83 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr4_+_150322151 | 0.80 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chrM_+_8603 | 0.79 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr12_-_83609217 | 0.78 |
ENSMUST00000222448.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr18_+_39126325 | 0.78 |
ENSMUST00000137497.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr18_+_39126178 | 0.78 |
ENSMUST00000097593.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr14_+_33662976 | 0.77 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr3_+_62327089 | 0.76 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr2_-_84545504 | 0.74 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr8_+_84728123 | 0.73 |
ENSMUST00000060357.15
ENSMUST00000239176.2 |
1700067K01Rik
|
RIKEN cDNA 1700067K01 gene |
| chr10_+_50770836 | 0.73 |
ENSMUST00000219436.2
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chrM_+_7006 | 0.73 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr17_+_3447465 | 0.72 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr12_-_56660054 | 0.72 |
ENSMUST00000072631.6
|
Nkx2-9
|
NK2 homeobox 9 |
| chr11_-_99213769 | 0.72 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr12_-_108145469 | 0.68 |
ENSMUST00000125916.3
ENSMUST00000109879.8 |
Setd3
|
SET domain containing 3 |
| chr18_+_56565188 | 0.66 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr16_-_45544960 | 0.66 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr4_+_109835224 | 0.63 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr3_-_92031247 | 0.62 |
ENSMUST00000070284.4
|
Prr9
|
proline rich 9 |
| chr11_+_114741948 | 0.62 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr9_+_78355474 | 0.61 |
ENSMUST00000034896.13
|
Mto1
|
mitochondrial tRNA translation optimization 1 |
| chr7_+_141503411 | 0.61 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr16_-_35891739 | 0.60 |
ENSMUST00000231351.2
ENSMUST00000004057.9 |
Fam162a
|
family with sequence similarity 162, member A |
| chr1_+_131455635 | 0.58 |
ENSMUST00000068613.5
|
Fam72a
|
family with sequence similarity 72, member A |
| chr5_+_36641922 | 0.57 |
ENSMUST00000060100.3
|
Ccdc96
|
coiled-coil domain containing 96 |
| chr16_+_37909363 | 0.57 |
ENSMUST00000023507.13
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr6_+_37847721 | 0.56 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr10_-_40018243 | 0.55 |
ENSMUST00000092566.8
ENSMUST00000213488.2 |
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
| chr11_+_116734104 | 0.54 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr14_-_70666513 | 0.54 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr5_+_14075281 | 0.54 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr2_+_22959452 | 0.53 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr10_+_39488930 | 0.53 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr15_+_100232810 | 0.53 |
ENSMUST00000075420.6
|
Mettl7a3
|
methyltransferase like 7A3 |
| chr5_+_141227245 | 0.53 |
ENSMUST00000085774.11
|
Sdk1
|
sidekick cell adhesion molecule 1 |
| chr4_-_63072367 | 0.52 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr19_+_6096606 | 0.52 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr8_-_84321069 | 0.51 |
ENSMUST00000019382.17
ENSMUST00000212630.2 ENSMUST00000165740.9 |
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr15_+_80139371 | 0.51 |
ENSMUST00000109605.5
ENSMUST00000229828.2 |
Atf4
|
activating transcription factor 4 |
| chr15_-_101801351 | 0.51 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr8_+_94763826 | 0.50 |
ENSMUST00000109556.9
ENSMUST00000093301.9 ENSMUST00000060632.8 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr5_-_74692327 | 0.49 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr15_+_100262423 | 0.49 |
ENSMUST00000175683.8
ENSMUST00000177211.2 |
Higd1c
|
HIG1 domain family, member 1C |
| chr6_-_87827993 | 0.49 |
ENSMUST00000204890.3
ENSMUST00000113617.3 ENSMUST00000113619.8 ENSMUST00000204653.3 ENSMUST00000032138.15 |
Cnbp
|
cellular nucleic acid binding protein |
| chr5_-_106844685 | 0.48 |
ENSMUST00000127434.8
ENSMUST00000112696.8 ENSMUST00000112698.8 |
Zfp644
|
zinc finger protein 644 |
| chr11_+_114742331 | 0.48 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_27055245 | 0.48 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr14_-_36820304 | 0.48 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr4_+_150938376 | 0.47 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr15_+_39522905 | 0.45 |
ENSMUST00000226410.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_14621669 | 0.45 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr10_-_8632519 | 0.45 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr4_-_14621805 | 0.45 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_+_49956441 | 0.44 |
ENSMUST00000112712.10
ENSMUST00000128451.8 ENSMUST00000053208.14 |
Lypd6
|
LY6/PLAUR domain containing 6 |
| chr12_-_104439589 | 0.44 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr12_+_76450941 | 0.42 |
ENSMUST00000080449.7
|
Hspa2
|
heat shock protein 2 |
| chr4_+_116078787 | 0.42 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr12_-_85197985 | 0.41 |
ENSMUST00000019379.9
ENSMUST00000221972.2 |
Rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr14_-_63221950 | 0.41 |
ENSMUST00000100493.3
|
Defb48
|
defensin beta 48 |
| chr8_-_84321032 | 0.41 |
ENSMUST00000163837.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr11_-_118103540 | 0.41 |
ENSMUST00000106302.9
ENSMUST00000151165.2 |
Cyth1
|
cytohesin 1 |
| chr4_-_14621497 | 0.40 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr19_-_6919755 | 0.40 |
ENSMUST00000099782.10
|
Gpr137
|
G protein-coupled receptor 137 |
| chr9_+_103940575 | 0.40 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr5_-_106844396 | 0.39 |
ENSMUST00000137285.8
ENSMUST00000124263.2 ENSMUST00000112695.4 ENSMUST00000155495.8 ENSMUST00000135108.2 ENSMUST00000149128.3 |
Zfp644
Gm28039
|
zinc finger protein 644 predicted gene, 28039 |
| chr3_+_103739877 | 0.39 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chrX_+_128650486 | 0.39 |
ENSMUST00000167619.9
ENSMUST00000037854.15 |
Diaph2
|
diaphanous related formin 2 |
| chr5_+_88731386 | 0.39 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr10_+_73657753 | 0.39 |
ENSMUST00000134009.9
ENSMUST00000177420.7 ENSMUST00000125006.9 |
Pcdh15
|
protocadherin 15 |
| chr13_+_34918820 | 0.38 |
ENSMUST00000039605.8
|
Fam50b
|
family with sequence similarity 50, member B |
| chr3_+_96175970 | 0.38 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chrM_+_7758 | 0.38 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr7_+_63835285 | 0.38 |
ENSMUST00000206263.2
ENSMUST00000206107.2 ENSMUST00000205731.2 ENSMUST00000206706.2 ENSMUST00000205690.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr2_-_122016670 | 0.38 |
ENSMUST00000028665.5
|
Patl2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr5_-_143279378 | 0.38 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr12_+_84498196 | 0.37 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr5_-_146158201 | 0.37 |
ENSMUST00000161574.8
|
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr2_+_177760959 | 0.37 |
ENSMUST00000108916.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr7_+_63835154 | 0.36 |
ENSMUST00000177102.8
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr16_-_22676264 | 0.35 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr10_-_107321938 | 0.35 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr1_-_92446141 | 0.35 |
ENSMUST00000189174.2
|
Olfr1414
|
olfactory receptor 1414 |
| chr12_+_72488625 | 0.35 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr11_-_99241924 | 0.35 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
| chr12_+_76451177 | 0.35 |
ENSMUST00000219555.2
|
Hspa2
|
heat shock protein 2 |
| chr18_+_24737009 | 0.34 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr15_-_101482320 | 0.34 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr9_+_32027335 | 0.34 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr3_-_141687987 | 0.34 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr2_+_177760768 | 0.34 |
ENSMUST00000108917.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr14_+_67953584 | 0.33 |
ENSMUST00000145542.8
ENSMUST00000125212.2 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr3_-_49711765 | 0.33 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr1_+_104696235 | 0.32 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr2_+_9887427 | 0.32 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr6_-_41752111 | 0.31 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chrX_-_111315519 | 0.31 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr18_+_37433852 | 0.31 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr6_+_8948608 | 0.31 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr1_+_161322219 | 0.31 |
ENSMUST00000086084.2
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr14_+_58308004 | 0.31 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr8_+_94879235 | 0.31 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr3_+_75464837 | 0.30 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr11_-_73290321 | 0.30 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr14_+_53698556 | 0.30 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr8_+_106877025 | 0.30 |
ENSMUST00000212963.2
ENSMUST00000034377.8 |
Pla2g15
|
phospholipase A2, group XV |
| chr19_+_25649767 | 0.30 |
ENSMUST00000053068.7
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr5_-_146158235 | 0.30 |
ENSMUST00000161859.8
|
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chrM_+_7779 | 0.29 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr11_-_69127848 | 0.29 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr17_+_34544632 | 0.29 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr15_+_102314578 | 0.29 |
ENSMUST00000170884.8
ENSMUST00000163709.8 |
Sp1
|
trans-acting transcription factor 1 |
| chr9_+_107440484 | 0.29 |
ENSMUST00000193418.2
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr11_+_95915366 | 0.28 |
ENSMUST00000103157.10
|
Gip
|
gastric inhibitory polypeptide |
| chr7_-_4909515 | 0.28 |
ENSMUST00000210663.2
|
Gm36210
|
predicted gene, 36210 |
| chr3_-_54962458 | 0.28 |
ENSMUST00000199352.2
ENSMUST00000198320.5 ENSMUST00000029368.7 |
Ccna1
|
cyclin A1 |
| chr17_-_37409147 | 0.28 |
ENSMUST00000216376.2
ENSMUST00000217372.2 |
Olfr91
|
olfactory receptor 91 |
| chr5_+_88731366 | 0.28 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr2_-_87322762 | 0.27 |
ENSMUST00000026957.4
|
Pramel7
|
PRAME like 7 |
| chr1_-_92446383 | 0.27 |
ENSMUST00000062353.12
|
Olfr1414
|
olfactory receptor 1414 |
| chr9_+_56858162 | 0.27 |
ENSMUST00000068856.5
|
Snupn
|
snurportin 1 |
| chr6_+_28480336 | 0.27 |
ENSMUST00000001460.14
ENSMUST00000167201.2 |
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr13_+_23758555 | 0.27 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr7_-_10488291 | 0.26 |
ENSMUST00000226874.2
ENSMUST00000227003.2 ENSMUST00000228561.2 ENSMUST00000228248.2 ENSMUST00000228526.2 ENSMUST00000228098.2 ENSMUST00000227940.2 ENSMUST00000228374.2 ENSMUST00000227702.2 |
Vmn1r71
|
vomeronasal 1 receptor 71 |
| chr2_+_153780138 | 0.26 |
ENSMUST00000109757.8
ENSMUST00000154281.3 |
Bpifb4
|
BPI fold containing family B, member 4 |
| chrM_+_2743 | 0.26 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr5_-_36641456 | 0.26 |
ENSMUST00000119916.2
ENSMUST00000031097.8 |
Tada2b
|
transcriptional adaptor 2B |
| chr15_+_82230155 | 0.25 |
ENSMUST00000023086.15
|
Smdt1
|
single-pass membrane protein with aspartate rich tail 1 |
| chr2_+_90229027 | 0.25 |
ENSMUST00000216111.3
|
Olfr1274
|
olfactory receptor 1274 |
| chr5_-_131645437 | 0.25 |
ENSMUST00000161804.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr15_-_76127600 | 0.25 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr3_-_32546380 | 0.25 |
ENSMUST00000164954.3
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr3_+_103740056 | 0.25 |
ENSMUST00000106822.2
|
Bcl2l15
|
BCLl2-like 15 |
| chr10_-_44024843 | 0.25 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr2_+_20742115 | 0.25 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr17_-_36773221 | 0.25 |
ENSMUST00000169950.2
ENSMUST00000057502.14 |
H2-M10.4
|
histocompatibility 2, M region locus 10.4 |
| chr7_-_25454177 | 0.24 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr3_-_129518723 | 0.24 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr19_+_3901797 | 0.24 |
ENSMUST00000072055.13
|
Chka
|
choline kinase alpha |
| chr14_+_32321341 | 0.24 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr9_-_96513529 | 0.24 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr10_+_73657689 | 0.24 |
ENSMUST00000064562.14
ENSMUST00000193174.6 ENSMUST00000105426.10 ENSMUST00000129404.9 ENSMUST00000131321.9 ENSMUST00000126920.9 ENSMUST00000147189.9 ENSMUST00000105424.10 ENSMUST00000092420.13 ENSMUST00000105429.10 ENSMUST00000193361.6 ENSMUST00000131724.9 ENSMUST00000152655.9 ENSMUST00000151116.9 ENSMUST00000155701.9 ENSMUST00000152819.9 ENSMUST00000125517.9 ENSMUST00000124046.8 ENSMUST00000146682.8 ENSMUST00000177107.8 ENSMUST00000149977.9 ENSMUST00000191854.6 |
Pcdh15
|
protocadherin 15 |
| chr11_+_102175985 | 0.24 |
ENSMUST00000156326.2
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr4_+_42949814 | 0.23 |
ENSMUST00000037872.10
ENSMUST00000098112.9 |
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr2_-_90054837 | 0.22 |
ENSMUST00000213994.3
|
Olfr1506
|
olfactory receptor 1506 |
| chr9_-_96601574 | 0.22 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr2_-_17465410 | 0.22 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr8_-_32440071 | 0.21 |
ENSMUST00000207678.3
|
Nrg1
|
neuregulin 1 |
| chr6_-_30304512 | 0.21 |
ENSMUST00000094543.3
ENSMUST00000102993.10 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr7_+_126549692 | 0.21 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chrX_+_159551009 | 0.20 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr4_+_136038301 | 0.20 |
ENSMUST00000084219.12
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr1_-_92412835 | 0.20 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chrX_+_56257374 | 0.20 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr3_-_49711706 | 0.20 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chrX_-_156198282 | 0.20 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_-_43840201 | 0.20 |
ENSMUST00000214281.2
|
Olfr157
|
olfactory receptor 157 |
| chr1_+_92545510 | 0.19 |
ENSMUST00000213247.2
|
Olfr12
|
olfactory receptor 12 |
| chr18_+_23548192 | 0.19 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr10_+_128173603 | 0.19 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr5_-_146157711 | 0.19 |
ENSMUST00000169407.9
ENSMUST00000161331.8 ENSMUST00000159074.3 ENSMUST00000067837.10 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr7_-_12096691 | 0.19 |
ENSMUST00000086228.3
|
Vmn1r84
|
vomeronasal 1 receptor 84 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.8 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.6 | 2.3 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.3 | 0.9 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 1.8 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.2 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.8 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.2 | 0.6 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.2 | 2.0 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 0.5 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.2 | 2.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 0.6 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 1.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 2.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.5 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.5 | GO:0000239 | pachytene(GO:0000239) |
| 0.1 | 2.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 1.4 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.1 | 0.3 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 0.3 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.3 | GO:0052564 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.1 | 0.4 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.3 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.3 | GO:0051030 | RNA import into nucleus(GO:0006404) snRNA transport(GO:0051030) |
| 0.1 | 0.5 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.5 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 1.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 2.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.6 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.0 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.9 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 3.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.8 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.9 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.8 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.5 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.0 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.8 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.2 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.6 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.6 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 2.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.3 | 0.8 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.3 | 2.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 8.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.7 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.4 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.1 | 0.8 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.5 | GO:1990037 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.3 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 6.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 9.4 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 1.1 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.3 | 2.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 5.2 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.2 | 6.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 4.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 2.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.5 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 1.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.5 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 3.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 1.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 2.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.8 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 2.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |