Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Ep300
Z-value: 0.79
Transcription factors associated with Ep300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ep300
|
ENSMUSG00000055024.13 | Ep300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ep300 | mm39_v1_chr15_+_81469538_81469598 | -0.17 | 3.3e-01 | Click! |
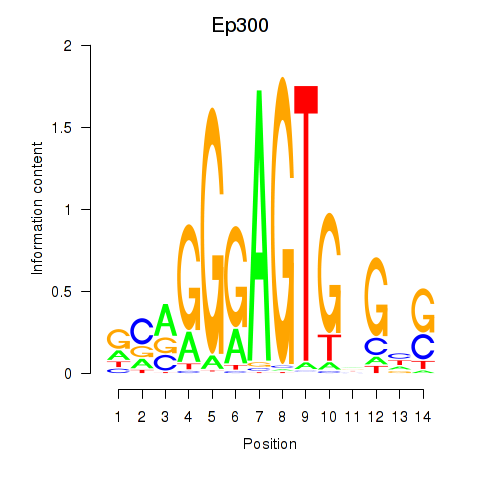
Activity profile of Ep300 motif
Sorted Z-values of Ep300 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ep300
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_133291302 | 1.85 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr19_+_44977512 | 1.60 |
ENSMUST00000026225.15
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr11_+_98239230 | 1.57 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr2_+_102489558 | 1.38 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_+_102488985 | 1.26 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr8_-_85526972 | 1.22 |
ENSMUST00000099070.10
|
Nfix
|
nuclear factor I/X |
| chr11_+_114742331 | 1.09 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_114741948 | 1.08 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr12_+_78273356 | 1.03 |
ENSMUST00000110388.10
|
Gphn
|
gephyrin |
| chr6_-_146403410 | 0.99 |
ENSMUST00000053273.15
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr17_+_35780977 | 0.95 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr1_-_60606237 | 0.91 |
ENSMUST00000142258.3
|
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr18_+_35963353 | 0.91 |
ENSMUST00000235169.2
|
Cxxc5
|
CXXC finger 5 |
| chr15_-_83054698 | 0.87 |
ENSMUST00000162178.8
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr13_-_119545520 | 0.85 |
ENSMUST00000069902.13
ENSMUST00000099149.10 ENSMUST00000109204.8 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr4_+_148123554 | 0.82 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr1_-_60605867 | 0.82 |
ENSMUST00000027168.12
ENSMUST00000090293.11 ENSMUST00000140485.8 |
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr7_-_140462187 | 0.80 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr15_-_75881289 | 0.80 |
ENSMUST00000170153.2
|
Fam83h
|
family with sequence similarity 83, member H |
| chr2_-_147887810 | 0.80 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chr7_-_140462221 | 0.79 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr11_-_120042019 | 0.79 |
ENSMUST00000179094.8
ENSMUST00000103018.11 ENSMUST00000045402.14 ENSMUST00000076697.13 ENSMUST00000053692.9 |
Slc38a10
|
solute carrier family 38, member 10 |
| chr12_-_44257109 | 0.77 |
ENSMUST00000015049.5
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr12_-_44256843 | 0.77 |
ENSMUST00000220421.2
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr13_-_119545479 | 0.76 |
ENSMUST00000223268.2
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr11_+_77405933 | 0.76 |
ENSMUST00000094004.5
|
Abhd15
|
abhydrolase domain containing 15 |
| chr13_-_55510595 | 0.75 |
ENSMUST00000021940.8
|
Lman2
|
lectin, mannose-binding 2 |
| chr2_-_26012751 | 0.73 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chrX_-_154121454 | 0.73 |
ENSMUST00000026328.11
|
Prdx4
|
peroxiredoxin 4 |
| chr9_+_55116209 | 0.71 |
ENSMUST00000034859.15
|
Fbxo22
|
F-box protein 22 |
| chr11_-_116197478 | 0.71 |
ENSMUST00000126731.8
|
Exoc7
|
exocyst complex component 7 |
| chr11_+_53660834 | 0.68 |
ENSMUST00000108920.10
ENSMUST00000140866.9 ENSMUST00000108922.9 |
Irf1
|
interferon regulatory factor 1 |
| chr9_+_55116474 | 0.68 |
ENSMUST00000146201.8
|
Fbxo22
|
F-box protein 22 |
| chr1_-_87501548 | 0.66 |
ENSMUST00000068681.12
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr4_-_149569614 | 0.65 |
ENSMUST00000126896.2
ENSMUST00000105693.2 ENSMUST00000030845.13 |
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr15_-_83054369 | 0.65 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr3_-_63872079 | 0.63 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr15_+_75468473 | 0.62 |
ENSMUST00000189944.7
ENSMUST00000023243.11 |
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr12_+_78273144 | 0.61 |
ENSMUST00000052472.6
|
Gphn
|
gephyrin |
| chr4_+_148123490 | 0.60 |
ENSMUST00000097788.11
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr15_+_75468494 | 0.55 |
ENSMUST00000189874.2
|
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr6_-_83098255 | 0.53 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chr7_+_127475968 | 0.52 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr2_-_102230602 | 0.52 |
ENSMUST00000152929.2
|
Trim44
|
tripartite motif-containing 44 |
| chr4_-_148123223 | 0.52 |
ENSMUST00000030879.12
ENSMUST00000137724.8 |
Clcn6
|
chloride channel, voltage-sensitive 6 |
| chrX_+_35375751 | 0.52 |
ENSMUST00000033418.8
|
Il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr3_-_90340910 | 0.51 |
ENSMUST00000196530.2
|
Ints3
|
integrator complex subunit 3 |
| chr11_-_116197523 | 0.50 |
ENSMUST00000133468.2
ENSMUST00000106411.10 ENSMUST00000106413.10 ENSMUST00000021147.14 |
Exoc7
|
exocyst complex component 7 |
| chr11_+_53661251 | 0.49 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr8_+_86567600 | 0.49 |
ENSMUST00000053771.14
ENSMUST00000161850.8 |
Phkb
|
phosphorylase kinase beta |
| chr1_-_183078488 | 0.49 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr9_+_44966464 | 0.48 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr7_-_139616234 | 0.47 |
ENSMUST00000209574.2
|
Tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr4_+_122889828 | 0.47 |
ENSMUST00000030407.8
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr4_+_122889737 | 0.47 |
ENSMUST00000106252.9
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr13_+_73752125 | 0.46 |
ENSMUST00000022102.9
|
Clptm1l
|
CLPTM1-like |
| chr14_-_32907446 | 0.45 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr18_-_68433265 | 0.43 |
ENSMUST00000152193.2
|
Fam210a
|
family with sequence similarity 210, member A |
| chr5_-_36987917 | 0.43 |
ENSMUST00000031002.10
|
Man2b2
|
mannosidase 2, alpha B2 |
| chr11_-_113456568 | 0.43 |
ENSMUST00000071539.10
ENSMUST00000106633.10 ENSMUST00000042657.16 ENSMUST00000149034.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr3_-_63872189 | 0.42 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr8_-_85414220 | 0.42 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr2_+_121001683 | 0.42 |
ENSMUST00000186659.7
ENSMUST00000039541.12 ENSMUST00000110657.2 |
Tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr6_-_83483868 | 0.42 |
ENSMUST00000014698.10
ENSMUST00000113888.3 |
Dguok
|
deoxyguanosine kinase |
| chr17_+_36152559 | 0.41 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr8_+_106877025 | 0.41 |
ENSMUST00000212963.2
ENSMUST00000034377.8 |
Pla2g15
|
phospholipase A2, group XV |
| chr6_-_39534765 | 0.40 |
ENSMUST00000036877.10
ENSMUST00000154149.2 |
Dennd2a
|
DENN/MADD domain containing 2A |
| chr6_-_113508536 | 0.40 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr3_-_101831729 | 0.40 |
ENSMUST00000190824.7
|
Slc22a15
|
solute carrier family 22 (organic anion/cation transporter), member 15 |
| chr8_-_11600689 | 0.40 |
ENSMUST00000049461.7
|
Cars2
|
cysteinyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr12_-_80306865 | 0.39 |
ENSMUST00000167327.2
|
Actn1
|
actinin, alpha 1 |
| chr17_+_87061117 | 0.39 |
ENSMUST00000024954.11
|
Epas1
|
endothelial PAS domain protein 1 |
| chr6_+_86415342 | 0.39 |
ENSMUST00000050497.14
ENSMUST00000203568.3 |
C87436
|
expressed sequence C87436 |
| chr9_+_105520154 | 0.39 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr6_-_149003003 | 0.38 |
ENSMUST00000127727.2
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr8_-_86567506 | 0.38 |
ENSMUST00000034140.9
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr1_+_87501721 | 0.38 |
ENSMUST00000166259.8
ENSMUST00000172222.8 ENSMUST00000163606.8 |
Neu2
|
neuraminidase 2 |
| chr16_-_94327689 | 0.37 |
ENSMUST00000023615.7
|
Vps26c
|
VPS26 endosomal protein sorting factor C |
| chr11_+_76070483 | 0.37 |
ENSMUST00000129853.8
|
Tlcd3a
|
TLC domain containing 3A |
| chr3_+_116306719 | 0.37 |
ENSMUST00000000349.11
ENSMUST00000197201.5 |
Dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr12_-_57592907 | 0.35 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr2_+_120294046 | 0.35 |
ENSMUST00000028749.15
ENSMUST00000110721.9 ENSMUST00000239364.2 |
Capn3
|
calpain 3 |
| chr10_-_83484467 | 0.35 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr4_-_133066594 | 0.35 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr2_-_90900628 | 0.34 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr4_-_133066549 | 0.34 |
ENSMUST00000105906.2
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr8_-_85413707 | 0.34 |
ENSMUST00000238301.2
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr10_-_59057570 | 0.34 |
ENSMUST00000220156.2
ENSMUST00000165971.3 |
Septin10
|
septin 10 |
| chr4_+_150321659 | 0.33 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr4_-_149569659 | 0.33 |
ENSMUST00000119921.8
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr10_-_57408512 | 0.33 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr8_+_45388466 | 0.33 |
ENSMUST00000191428.7
|
Fat1
|
FAT atypical cadherin 1 |
| chr5_-_142891686 | 0.33 |
ENSMUST00000106216.3
|
Actb
|
actin, beta |
| chr8_-_70686746 | 0.33 |
ENSMUST00000130319.2
|
Armc6
|
armadillo repeat containing 6 |
| chr2_-_155571279 | 0.33 |
ENSMUST00000040833.5
|
Edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr14_-_54647647 | 0.32 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr10_+_59057767 | 0.32 |
ENSMUST00000182161.2
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr3_-_90340830 | 0.31 |
ENSMUST00000029542.12
|
Ints3
|
integrator complex subunit 3 |
| chr6_-_149003171 | 0.31 |
ENSMUST00000111557.8
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr9_-_79700660 | 0.31 |
ENSMUST00000034878.12
|
Tmem30a
|
transmembrane protein 30A |
| chr4_-_118266416 | 0.31 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr2_-_102231208 | 0.31 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr19_-_46136765 | 0.31 |
ENSMUST00000026259.16
|
Pitx3
|
paired-like homeodomain transcription factor 3 |
| chr14_+_21549798 | 0.30 |
ENSMUST00000182855.8
ENSMUST00000182405.9 ENSMUST00000069648.14 |
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr2_+_121001636 | 0.29 |
ENSMUST00000110658.8
|
Tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr9_-_20887967 | 0.29 |
ENSMUST00000214218.2
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr3_+_88857929 | 0.28 |
ENSMUST00000186583.7
|
Ash1l
|
ASH1 like histone lysine methyltransferase |
| chr11_-_93846453 | 0.28 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr6_+_86415648 | 0.28 |
ENSMUST00000113700.8
|
C87436
|
expressed sequence C87436 |
| chr7_+_89053562 | 0.28 |
ENSMUST00000058755.5
|
Fzd4
|
frizzled class receptor 4 |
| chr19_+_44994905 | 0.27 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
| chr7_+_44805280 | 0.27 |
ENSMUST00000107811.4
ENSMUST00000211414.2 |
Pih1d1
|
PIH1 domain containing 1 |
| chrX_+_60753074 | 0.27 |
ENSMUST00000075983.6
|
Ldoc1
|
regulator of NFKB signaling |
| chr5_-_142891565 | 0.27 |
ENSMUST00000171419.8
|
Actb
|
actin, beta |
| chr15_-_97665679 | 0.27 |
ENSMUST00000129223.9
ENSMUST00000126854.9 ENSMUST00000135080.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr17_-_32503107 | 0.26 |
ENSMUST00000237692.2
|
Brd4
|
bromodomain containing 4 |
| chr15_+_76215431 | 0.26 |
ENSMUST00000023221.13
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr17_-_24863956 | 0.26 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr4_+_133246274 | 0.25 |
ENSMUST00000149807.2
ENSMUST00000042919.16 ENSMUST00000153811.2 ENSMUST00000105901.2 ENSMUST00000121797.2 |
Kdf1
|
keratinocyte differentiation factor 1 |
| chr9_-_57513510 | 0.25 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chrX_+_106860083 | 0.25 |
ENSMUST00000143975.8
ENSMUST00000144695.8 ENSMUST00000167154.2 |
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr4_+_59805829 | 0.25 |
ENSMUST00000030080.7
|
Snx30
|
sorting nexin family member 30 |
| chr17_-_24863907 | 0.25 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr3_+_88461059 | 0.25 |
ENSMUST00000008748.8
|
Ubqln4
|
ubiquilin 4 |
| chr15_+_75881712 | 0.25 |
ENSMUST00000187868.3
|
Iqank1
|
IQ motif and ankyrin repeat containing 1 |
| chr9_-_79700789 | 0.25 |
ENSMUST00000120690.2
|
Tmem30a
|
transmembrane protein 30A |
| chrX_+_57075981 | 0.24 |
ENSMUST00000088627.11
|
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr9_+_6168638 | 0.24 |
ENSMUST00000214892.2
|
Pdgfd
|
platelet-derived growth factor, D polypeptide |
| chr2_+_30282414 | 0.24 |
ENSMUST00000123202.8
ENSMUST00000113612.10 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr9_-_20888054 | 0.23 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr8_-_122556219 | 0.23 |
ENSMUST00000174717.8
ENSMUST00000174192.2 |
Klhdc4
|
kelch domain containing 4 |
| chr8_-_122556258 | 0.23 |
ENSMUST00000045884.17
|
Klhdc4
|
kelch domain containing 4 |
| chr18_-_68433398 | 0.23 |
ENSMUST00000042852.7
|
Fam210a
|
family with sequence similarity 210, member A |
| chr7_+_65343156 | 0.22 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr11_-_76070325 | 0.22 |
ENSMUST00000167114.8
ENSMUST00000094015.11 ENSMUST00000108419.9 ENSMUST00000170730.3 ENSMUST00000129256.2 ENSMUST00000056601.11 |
Vps53
|
VPS53 GARP complex subunit |
| chr8_-_65302573 | 0.22 |
ENSMUST00000210166.2
|
Klhl2
|
kelch-like 2, Mayven |
| chr2_-_52566583 | 0.22 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr7_-_126294902 | 0.21 |
ENSMUST00000144897.2
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr6_+_86415698 | 0.21 |
ENSMUST00000113698.8
|
C87436
|
expressed sequence C87436 |
| chr9_-_110237276 | 0.21 |
ENSMUST00000040021.12
|
Ptpn23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr10_-_120735000 | 0.21 |
ENSMUST00000092143.12
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr3_-_88857578 | 0.20 |
ENSMUST00000174402.8
ENSMUST00000174077.8 |
Dap3
|
death associated protein 3 |
| chr4_+_150321142 | 0.20 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr7_-_139616167 | 0.20 |
ENSMUST00000026547.9
|
Tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr16_-_52272828 | 0.20 |
ENSMUST00000170035.8
ENSMUST00000164728.8 ENSMUST00000168071.2 |
Alcam
|
activated leukocyte cell adhesion molecule |
| chr5_+_65921414 | 0.20 |
ENSMUST00000201615.4
ENSMUST00000087264.4 |
N4bp2
|
NEDD4 binding protein 2 |
| chr2_-_155356716 | 0.19 |
ENSMUST00000029131.11
|
Ggt7
|
gamma-glutamyltransferase 7 |
| chr17_+_78507669 | 0.19 |
ENSMUST00000112498.3
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like) |
| chr10_-_40133558 | 0.19 |
ENSMUST00000216847.2
ENSMUST00000213628.2 ENSMUST00000217537.2 ENSMUST00000019982.9 |
Gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr11_+_49135018 | 0.19 |
ENSMUST00000167400.8
ENSMUST00000081794.7 |
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr18_+_24737009 | 0.19 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr15_-_97665524 | 0.19 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr8_+_104828253 | 0.19 |
ENSMUST00000034339.10
|
Cdh5
|
cadherin 5 |
| chr11_+_76092833 | 0.18 |
ENSMUST00000094014.10
|
Tlcd3a
|
TLC domain containing 3A |
| chr9_-_29323032 | 0.18 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr6_+_110622533 | 0.18 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr9_+_100956145 | 0.18 |
ENSMUST00000189616.2
|
Msl2
|
MSL complex subunit 2 |
| chrX_-_151820545 | 0.18 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr13_+_120151982 | 0.18 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr8_+_70686836 | 0.18 |
ENSMUST00000164403.8
ENSMUST00000093458.11 |
Sugp2
|
SURP and G patch domain containing 2 |
| chr2_+_24276545 | 0.18 |
ENSMUST00000127242.2
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr11_-_93859064 | 0.18 |
ENSMUST00000107844.3
ENSMUST00000170303.2 |
Nme1
Gm20390
|
NME/NM23 nucleoside diphosphate kinase 1 predicted gene 20390 |
| chr11_-_62348115 | 0.18 |
ENSMUST00000069456.11
ENSMUST00000018645.13 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr10_+_77095052 | 0.18 |
ENSMUST00000020493.9
|
Pofut2
|
protein O-fucosyltransferase 2 |
| chr11_+_76092909 | 0.17 |
ENSMUST00000169560.2
|
Tlcd3a
|
TLC domain containing 3A |
| chr9_+_21914513 | 0.17 |
ENSMUST00000215795.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr2_-_121001577 | 0.17 |
ENSMUST00000163766.8
ENSMUST00000146243.2 |
Zscan29
|
zinc finger SCAN domains 29 |
| chrX_+_119199956 | 0.17 |
ENSMUST00000113364.10
ENSMUST00000050239.16 ENSMUST00000113358.10 |
Pcdh11x
|
protocadherin 11 X-linked |
| chr14_+_66074751 | 0.16 |
ENSMUST00000022614.7
|
Ccdc25
|
coiled-coil domain containing 25 |
| chr3_+_90444537 | 0.16 |
ENSMUST00000098911.10
|
S100a16
|
S100 calcium binding protein A16 |
| chrX_+_57076359 | 0.16 |
ENSMUST00000088631.11
ENSMUST00000088629.4 |
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr2_-_90900525 | 0.16 |
ENSMUST00000153367.2
ENSMUST00000079976.10 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr11_-_76969230 | 0.15 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1 |
| chr3_-_88857707 | 0.15 |
ENSMUST00000090938.11
|
Dap3
|
death associated protein 3 |
| chr1_-_37469037 | 0.15 |
ENSMUST00000027286.7
|
Coa5
|
cytochrome C oxidase assembly factor 5 |
| chr9_-_121106209 | 0.14 |
ENSMUST00000051479.13
ENSMUST00000171923.8 |
Ulk4
|
unc-51-like kinase 4 |
| chr18_+_31937129 | 0.14 |
ENSMUST00000082319.15
ENSMUST00000234957.2 ENSMUST00000025264.8 ENSMUST00000234344.2 |
Wdr33
|
WD repeat domain 33 |
| chr16_+_48104098 | 0.14 |
ENSMUST00000096045.9
ENSMUST00000050705.4 |
Dppa4
|
developmental pluripotency associated 4 |
| chr2_+_163444214 | 0.14 |
ENSMUST00000171696.8
ENSMUST00000109408.10 |
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chrX_+_149830166 | 0.13 |
ENSMUST00000026296.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr1_+_75427080 | 0.13 |
ENSMUST00000113577.8
|
Asic4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr5_-_122752508 | 0.13 |
ENSMUST00000127220.8
ENSMUST00000031426.14 |
Ift81
|
intraflagellar transport 81 |
| chr1_+_171238911 | 0.13 |
ENSMUST00000160486.8
|
Usf1
|
upstream transcription factor 1 |
| chr19_+_23736205 | 0.12 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr5_-_53864595 | 0.12 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr3_+_90444613 | 0.12 |
ENSMUST00000107335.2
|
S100a16
|
S100 calcium binding protein A16 |
| chr3_+_138911648 | 0.12 |
ENSMUST00000062306.7
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr6_-_56546088 | 0.11 |
ENSMUST00000203372.3
|
Pde1c
|
phosphodiesterase 1C |
| chr10_+_79752797 | 0.11 |
ENSMUST00000045529.3
|
Kiss1r
|
KISS1 receptor |
| chr6_+_99669640 | 0.11 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr9_-_29323500 | 0.11 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr1_+_74811045 | 0.11 |
ENSMUST00000006716.8
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr15_+_6416079 | 0.11 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr14_-_25903524 | 0.10 |
ENSMUST00000052286.16
|
Plac9a
|
placenta specific 9a |
| chr11_+_5905693 | 0.10 |
ENSMUST00000002818.9
|
Ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chrX_-_106859842 | 0.10 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr2_+_179684288 | 0.10 |
ENSMUST00000041126.9
|
Ss18l1
|
SS18, nBAF chromatin remodeling complex subunit like 1 |
| chr11_-_116197994 | 0.10 |
ENSMUST00000124281.2
|
Exoc7
|
exocyst complex component 7 |
| chr3_-_51184730 | 0.09 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr15_+_8138805 | 0.09 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.5 | 1.6 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.4 | 1.2 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 0.4 | 2.6 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 1.4 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.3 | 0.8 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 0.7 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 1.2 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.2 | 0.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 1.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.5 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.2 | 1.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.7 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.4 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 1.6 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 0.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 1.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.5 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 0.3 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 0.3 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.1 | 0.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.4 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.4 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.1 | 0.3 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.3 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.7 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.0 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.4 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.2 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 1.2 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 | 0.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.3 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.4 | GO:0006517 | mannose metabolic process(GO:0006013) protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.7 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.3 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.0 | 0.1 | GO:0060166 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) olfactory pit development(GO:0060166) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 1.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.7 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.8 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 2.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 1.7 | GO:0048675 | axon extension(GO:0048675) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.5 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 0.4 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 1.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.7 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.4 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 2.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 1.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.5 | 1.6 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.5 | 1.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.5 | 1.8 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.4 | 2.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 1.2 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.2 | 1.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 1.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 0.6 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 1.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.4 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 1.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.2 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 0.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.7 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.4 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 1.6 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.7 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 5.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 3.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |