Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
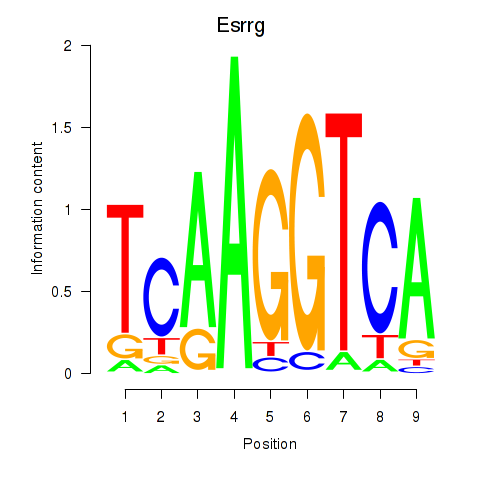
Results for Esrrg
Z-value: 0.82
Transcription factors associated with Esrrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrg
|
ENSMUSG00000026610.14 | Esrrg |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrrg | mm39_v1_chr1_+_187340952_187340988 | -0.06 | 7.4e-01 | Click! |
Activity profile of Esrrg motif
Sorted Z-values of Esrrg motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrg
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7943365 | 2.59 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr4_-_107164347 | 1.40 |
ENSMUST00000082426.11
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr9_-_44891626 | 1.20 |
ENSMUST00000002101.12
ENSMUST00000160886.2 |
Cd3g
|
CD3 antigen, gamma polypeptide |
| chr4_+_34893772 | 1.20 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr17_-_35077089 | 1.15 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr19_-_43512929 | 1.12 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr4_-_107164315 | 1.07 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr2_-_73741664 | 0.95 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr16_+_32427738 | 0.90 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr17_-_32639936 | 0.77 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr17_-_26087696 | 0.73 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr16_-_4831349 | 0.70 |
ENSMUST00000201077.2
ENSMUST00000202281.4 ENSMUST00000090453.9 ENSMUST00000023191.17 |
Rogdi
|
rogdi homolog |
| chrX_-_135877607 | 0.68 |
ENSMUST00000127404.8
ENSMUST00000113071.2 |
Tmsb15l
Tmsb15b1
|
thymosin beta 15b like thymosin beta 15b1 |
| chr10_-_80691009 | 0.68 |
ENSMUST00000220225.2
ENSMUST00000035775.9 |
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_-_111638722 | 0.66 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr7_+_127728712 | 0.60 |
ENSMUST00000033053.8
ENSMUST00000205460.2 |
Itgax
|
integrin alpha X |
| chr4_+_140688514 | 0.58 |
ENSMUST00000010007.9
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr14_-_30637344 | 0.58 |
ENSMUST00000226547.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr9_+_50515207 | 0.57 |
ENSMUST00000044051.6
|
Timm8b
|
translocase of inner mitochondrial membrane 8B |
| chr11_-_53321606 | 0.56 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr19_-_47079052 | 0.53 |
ENSMUST00000235771.2
ENSMUST00000096014.5 ENSMUST00000236170.2 |
Atp5md
|
ATP synthase membrane subunit DAPIT |
| chr2_+_131028861 | 0.52 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr9_-_50515089 | 0.51 |
ENSMUST00000000175.6
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr19_+_4203603 | 0.50 |
ENSMUST00000236632.2
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr11_+_76297969 | 0.50 |
ENSMUST00000021203.7
ENSMUST00000152183.2 |
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr1_-_167221344 | 0.50 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr13_-_86194889 | 0.49 |
ENSMUST00000131011.2
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr19_+_6973511 | 0.48 |
ENSMUST00000088223.7
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr4_+_13751297 | 0.48 |
ENSMUST00000105566.9
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr19_-_6957789 | 0.47 |
ENSMUST00000070878.9
ENSMUST00000177752.9 |
Fkbp2
|
FK506 binding protein 2 |
| chr6_+_54249817 | 0.47 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr2_+_140012560 | 0.46 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr11_+_121593582 | 0.46 |
ENSMUST00000125580.2
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr4_-_45108038 | 0.45 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr3_+_146156220 | 0.44 |
ENSMUST00000061937.13
ENSMUST00000029840.4 |
Ctbs
|
chitobiase |
| chr14_+_25694594 | 0.44 |
ENSMUST00000022419.7
|
Ppif
|
peptidylprolyl isomerase F (cyclophilin F) |
| chr10_+_78410803 | 0.44 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr5_-_92496730 | 0.44 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr6_-_92160952 | 0.43 |
ENSMUST00000140438.2
|
Mrps25
|
mitochondrial ribosomal protein S25 |
| chr11_-_100986192 | 0.42 |
ENSMUST00000019447.15
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr8_-_84771610 | 0.42 |
ENSMUST00000061923.5
|
Rln3
|
relaxin 3 |
| chr2_+_109522781 | 0.42 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr9_-_63306497 | 0.40 |
ENSMUST00000168665.3
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chr3_+_146205864 | 0.40 |
ENSMUST00000119130.2
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr6_-_114018982 | 0.40 |
ENSMUST00000101045.10
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr7_-_100232276 | 0.39 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr10_+_79552421 | 0.39 |
ENSMUST00000099513.8
ENSMUST00000020581.3 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr6_-_71800788 | 0.38 |
ENSMUST00000065103.4
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr16_-_91728531 | 0.37 |
ENSMUST00000023677.10
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr19_-_4889284 | 0.36 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr7_-_105131407 | 0.36 |
ENSMUST00000047040.4
|
Cavin3
|
caveolae associated 3 |
| chr17_+_17669082 | 0.35 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr10_-_127016448 | 0.35 |
ENSMUST00000222911.3
ENSMUST00000095270.3 |
Slc26a10
|
solute carrier family 26, member 10 |
| chrX_+_74425990 | 0.34 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr19_-_4889314 | 0.33 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr5_-_24650164 | 0.33 |
ENSMUST00000115043.8
ENSMUST00000115041.2 ENSMUST00000030800.13 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr10_-_43416941 | 0.32 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr16_-_91728162 | 0.32 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr7_-_16761732 | 0.31 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr19_-_7218363 | 0.31 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr4_-_82423511 | 0.31 |
ENSMUST00000050872.15
ENSMUST00000064770.9 |
Nfib
|
nuclear factor I/B |
| chrX_-_7884688 | 0.30 |
ENSMUST00000033503.3
|
Glod5
|
glyoxalase domain containing 5 |
| chr11_-_52165682 | 0.30 |
ENSMUST00000238914.2
|
Tcf7
|
transcription factor 7, T cell specific |
| chr2_-_77000878 | 0.29 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr7_-_16761790 | 0.29 |
ENSMUST00000003183.12
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr9_-_121686601 | 0.29 |
ENSMUST00000213124.2
ENSMUST00000215300.2 ENSMUST00000213147.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr19_-_6973393 | 0.29 |
ENSMUST00000041686.10
ENSMUST00000180765.2 |
Nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr16_+_22769822 | 0.29 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr18_-_36877571 | 0.29 |
ENSMUST00000014438.5
|
Ndufa2
|
NADH:ubiquinone oxidoreductase subunit A2 |
| chr4_-_82423985 | 0.28 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr1_+_133249613 | 0.28 |
ENSMUST00000007433.5
|
Kiss1
|
KiSS-1 metastasis-suppressor |
| chr19_+_6449887 | 0.28 |
ENSMUST00000146601.8
ENSMUST00000150713.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr17_-_33879224 | 0.27 |
ENSMUST00000130946.8
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr1_-_175453117 | 0.27 |
ENSMUST00000027810.14
|
Fh1
|
fumarate hydratase 1 |
| chr15_-_102579463 | 0.27 |
ENSMUST00000185641.7
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) |
| chr8_-_70975813 | 0.27 |
ENSMUST00000121623.8
ENSMUST00000093456.12 ENSMUST00000118850.8 |
Kxd1
|
KxDL motif containing 1 |
| chr16_-_95260104 | 0.27 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr14_+_79086492 | 0.26 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr17_+_48047955 | 0.26 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr4_-_128856213 | 0.26 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr10_+_127919142 | 0.25 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr1_-_43235914 | 0.25 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2 |
| chr17_-_26004507 | 0.25 |
ENSMUST00000140738.8
ENSMUST00000145053.2 ENSMUST00000138759.8 ENSMUST00000133071.8 ENSMUST00000077938.10 |
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr7_-_4525426 | 0.25 |
ENSMUST00000209148.2
ENSMUST00000098859.10 |
Tnni3
|
troponin I, cardiac 3 |
| chr9_-_107546166 | 0.24 |
ENSMUST00000177567.8
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr9_-_15212849 | 0.24 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr18_+_77861656 | 0.24 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr7_+_28050077 | 0.24 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr6_+_70703409 | 0.24 |
ENSMUST00000103410.3
|
Igkc
|
immunoglobulin kappa constant |
| chr4_-_82423944 | 0.23 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr5_-_25200745 | 0.23 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr7_-_101899294 | 0.22 |
ENSMUST00000106923.2
ENSMUST00000098230.11 |
Rhog
|
ras homolog family member G |
| chr9_+_37313287 | 0.22 |
ENSMUST00000115048.10
ENSMUST00000115046.9 ENSMUST00000102895.7 ENSMUST00000239486.2 |
Robo4
|
roundabout guidance receptor 4 |
| chr7_+_83281167 | 0.21 |
ENSMUST00000075418.15
|
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chrX_-_47602395 | 0.20 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr11_+_98632631 | 0.20 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr15_+_76227695 | 0.20 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr17_-_26004298 | 0.20 |
ENSMUST00000150324.8
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr16_-_29363671 | 0.20 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr1_+_36730530 | 0.20 |
ENSMUST00000081180.7
ENSMUST00000193210.6 ENSMUST00000195151.6 |
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr7_-_37472979 | 0.19 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr10_+_80100812 | 0.19 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr7_-_4525793 | 0.19 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr9_-_43027809 | 0.19 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr6_+_91134358 | 0.18 |
ENSMUST00000155007.2
|
Hdac11
|
histone deacetylase 11 |
| chr19_+_6449776 | 0.18 |
ENSMUST00000113468.8
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr13_-_64645606 | 0.18 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr3_+_85481416 | 0.17 |
ENSMUST00000107672.8
ENSMUST00000127348.8 ENSMUST00000107674.2 |
Gatb
|
glutamyl-tRNA(Gln) amidotransferase, subunit B |
| chr1_+_36510670 | 0.17 |
ENSMUST00000153128.2
|
Cnnm4
|
cyclin M4 |
| chr11_-_94568228 | 0.17 |
ENSMUST00000116349.9
|
Xylt2
|
xylosyltransferase II |
| chr9_-_121686643 | 0.16 |
ENSMUST00000215007.2
ENSMUST00000216914.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr2_-_84508385 | 0.15 |
ENSMUST00000189772.2
ENSMUST00000053664.9 ENSMUST00000111664.8 |
Gm28635
Tmx2
|
predicted gene 28635 thioredoxin-related transmembrane protein 2 |
| chr2_+_121125918 | 0.15 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chr8_+_121395047 | 0.15 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr4_+_138161958 | 0.15 |
ENSMUST00000044058.11
ENSMUST00000105813.8 ENSMUST00000105815.2 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr11_-_100332619 | 0.15 |
ENSMUST00000107398.8
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr1_+_75377616 | 0.14 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr12_-_103597663 | 0.14 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr7_+_45224524 | 0.14 |
ENSMUST00000210811.2
|
Bcat2
|
branched chain aminotransferase 2, mitochondrial |
| chr8_+_95564949 | 0.14 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr17_+_69463846 | 0.14 |
ENSMUST00000238898.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr15_-_99355623 | 0.14 |
ENSMUST00000023747.14
|
Nckap5l
|
NCK-associated protein 5-like |
| chr12_-_113896002 | 0.14 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr6_-_124698805 | 0.13 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr5_-_121665249 | 0.13 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr9_+_107468146 | 0.13 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr15_-_35938155 | 0.13 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr2_-_155771938 | 0.13 |
ENSMUST00000152766.8
ENSMUST00000139232.8 ENSMUST00000109632.8 ENSMUST00000006036.13 ENSMUST00000142655.2 ENSMUST00000159238.2 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr9_-_106768601 | 0.12 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr17_-_56891576 | 0.12 |
ENSMUST00000075510.12
|
Safb2
|
scaffold attachment factor B2 |
| chr18_+_57275854 | 0.12 |
ENSMUST00000139892.2
|
Megf10
|
multiple EGF-like-domains 10 |
| chr9_+_37313193 | 0.12 |
ENSMUST00000214185.3
|
Robo4
|
roundabout guidance receptor 4 |
| chr11_-_115590133 | 0.12 |
ENSMUST00000106499.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr16_+_44979086 | 0.12 |
ENSMUST00000023343.4
|
Atg3
|
autophagy related 3 |
| chr6_+_83326424 | 0.11 |
ENSMUST00000136501.2
|
Bola3
|
bolA-like 3 (E. coli) |
| chr3_+_68776884 | 0.11 |
ENSMUST00000054551.3
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr11_+_70655035 | 0.11 |
ENSMUST00000060444.6
|
Zfp3
|
zinc finger protein 3 |
| chr7_-_110462446 | 0.11 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr3_-_94693780 | 0.11 |
ENSMUST00000107273.9
ENSMUST00000238849.2 |
Cgn
|
cingulin |
| chr17_+_84013575 | 0.11 |
ENSMUST00000112350.8
ENSMUST00000112349.9 ENSMUST00000112352.10 ENSMUST00000067826.15 |
Mta3
|
metastasis associated 3 |
| chr4_+_131600918 | 0.11 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr10_-_4338032 | 0.10 |
ENSMUST00000100078.10
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr3_+_32791139 | 0.10 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr4_-_40279382 | 0.10 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr7_-_37472290 | 0.10 |
ENSMUST00000176205.8
|
Zfp536
|
zinc finger protein 536 |
| chr12_+_104998895 | 0.09 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr10_-_84276454 | 0.09 |
ENSMUST00000020220.15
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr15_-_79169671 | 0.09 |
ENSMUST00000170955.2
ENSMUST00000165408.8 |
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr2_+_129854256 | 0.09 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr12_+_85645801 | 0.09 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr7_-_4448631 | 0.09 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr7_+_120234399 | 0.08 |
ENSMUST00000033176.7
ENSMUST00000208400.2 |
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr15_-_63932176 | 0.08 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr14_-_70680882 | 0.08 |
ENSMUST00000000793.13
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr15_-_11907267 | 0.08 |
ENSMUST00000228489.2
|
Npr3
|
natriuretic peptide receptor 3 |
| chr14_-_31807552 | 0.08 |
ENSMUST00000022461.11
ENSMUST00000067955.12 ENSMUST00000124303.9 |
Dph3
|
diphthamine biosynthesis 3 |
| chr13_-_21685588 | 0.08 |
ENSMUST00000044043.3
|
Cox5b-ps
|
cytochrome c oxidase subunit 5B, pseudogene |
| chr15_+_78314251 | 0.08 |
ENSMUST00000229622.2
ENSMUST00000162808.2 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr3_+_121243395 | 0.08 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr11_+_7013422 | 0.08 |
ENSMUST00000020706.5
|
Adcy1
|
adenylate cyclase 1 |
| chr12_+_85646162 | 0.08 |
ENSMUST00000050687.14
|
Jdp2
|
Jun dimerization protein 2 |
| chr9_+_65536892 | 0.08 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr17_+_6926452 | 0.08 |
ENSMUST00000097430.10
|
Sytl3
|
synaptotagmin-like 3 |
| chr7_-_134100659 | 0.08 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr11_+_42310557 | 0.07 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr2_+_164627743 | 0.07 |
ENSMUST00000174070.8
ENSMUST00000172577.8 ENSMUST00000056181.7 |
Snx21
|
sorting nexin family member 21 |
| chr3_-_32791296 | 0.07 |
ENSMUST00000043966.8
|
Mrpl47
|
mitochondrial ribosomal protein L47 |
| chrX_-_104918911 | 0.07 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr19_+_13608985 | 0.07 |
ENSMUST00000217182.3
|
Olfr1489
|
olfactory receptor 1489 |
| chr16_+_4501934 | 0.06 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr11_+_120252360 | 0.06 |
ENSMUST00000026445.3
|
Fscn2
|
fascin actin-bundling protein 2 |
| chr16_-_20245138 | 0.06 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr17_-_45785752 | 0.06 |
ENSMUST00000233553.2
ENSMUST00000233769.2 ENSMUST00000024731.9 |
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr2_+_25132941 | 0.06 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr13_+_76115824 | 0.05 |
ENSMUST00000022081.3
|
Spata9
|
spermatogenesis associated 9 |
| chr6_-_113508536 | 0.05 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr12_-_110945376 | 0.05 |
ENSMUST00000142012.2
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr8_+_85696396 | 0.05 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr11_+_68979308 | 0.05 |
ENSMUST00000021273.13
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chrX_+_56008685 | 0.05 |
ENSMUST00000096431.10
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr13_-_91372072 | 0.05 |
ENSMUST00000022119.6
|
Atg10
|
autophagy related 10 |
| chr17_+_29251602 | 0.05 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr11_+_74540284 | 0.05 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr1_-_16589425 | 0.05 |
ENSMUST00000159558.8
ENSMUST00000054668.13 ENSMUST00000162627.8 ENSMUST00000162007.8 ENSMUST00000128957.9 ENSMUST00000115359.10 ENSMUST00000151888.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr19_+_6450641 | 0.04 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr14_+_58313964 | 0.04 |
ENSMUST00000166770.2
|
Fgf9
|
fibroblast growth factor 9 |
| chr8_+_85696695 | 0.04 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr5_+_145141333 | 0.04 |
ENSMUST00000085671.10
ENSMUST00000031601.8 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr11_-_115590318 | 0.04 |
ENSMUST00000106497.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr7_-_115837034 | 0.04 |
ENSMUST00000182834.8
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr15_-_99355593 | 0.04 |
ENSMUST00000161948.2
|
Nckap5l
|
NCK-associated protein 5-like |
| chr15_+_30173197 | 0.04 |
ENSMUST00000226119.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr18_-_43610829 | 0.03 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr16_-_20245071 | 0.03 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr5_+_129802127 | 0.03 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr19_+_6450553 | 0.03 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr8_-_70975734 | 0.03 |
ENSMUST00000137610.3
|
Kxd1
|
KxDL motif containing 1 |
| chr2_+_145627900 | 0.03 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr2_+_32477069 | 0.03 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr17_-_27247581 | 0.03 |
ENSMUST00000143158.3
|
Bak1
|
BCL2-antagonist/killer 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.2 | 0.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 1.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 2.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.5 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 0.4 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.8 | GO:0032824 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.6 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.3 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 0.4 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 0.1 | 0.4 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 1.2 | GO:0032275 | luteinizing hormone secretion(GO:0032275) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.6 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 1.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.2 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 0.3 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.3 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.3 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.4 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.4 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.3 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.0 | 0.5 | GO:2000507 | positive regulation of brown fat cell differentiation(GO:0090336) positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 2.5 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.2 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.5 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.0 | 1.2 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.8 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.6 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.0 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.4 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 1.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.4 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.1 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 1.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.4 | 1.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 0.5 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.2 | 2.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.8 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 2.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 3.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |