Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Etv6
Z-value: 0.93
Transcription factors associated with Etv6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv6
|
ENSMUSG00000030199.17 | Etv6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv6 | mm39_v1_chr6_+_134012602_134012683 | -0.41 | 1.2e-02 | Click! |
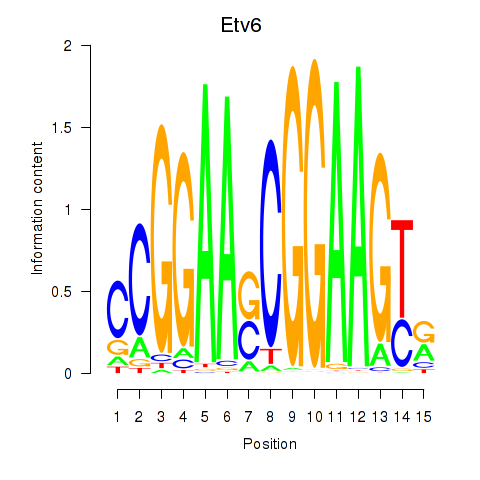
Activity profile of Etv6 motif
Sorted Z-values of Etv6 motif
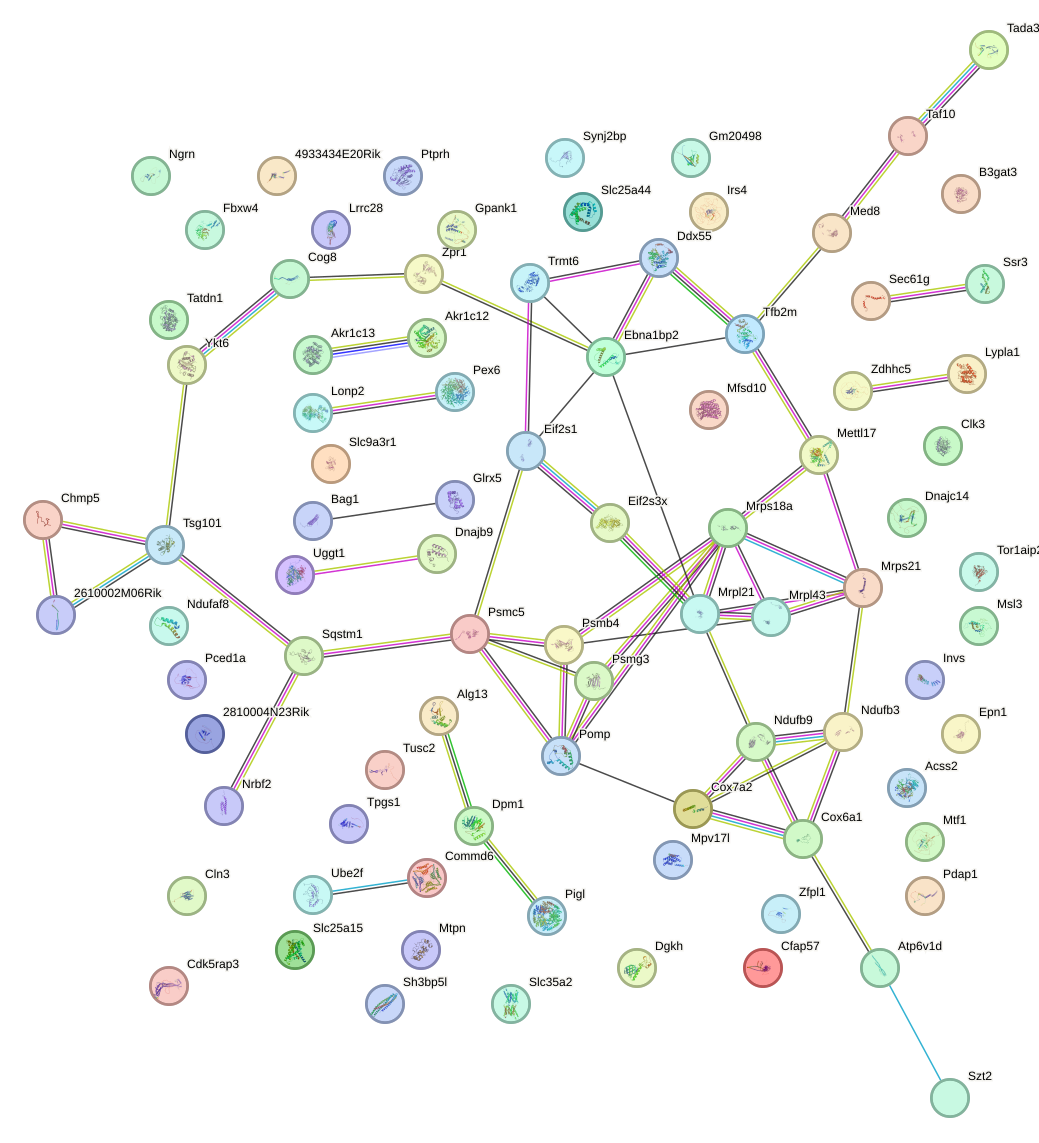
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_155359868 | 1.69 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr4_+_118266526 | 1.60 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr2_+_155360015 | 1.57 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr4_+_118266582 | 1.49 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr5_-_149559667 | 1.07 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr5_-_149559792 | 1.07 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr5_-_149559636 | 0.87 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_+_167136217 | 0.86 |
ENSMUST00000193446.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr14_+_52122299 | 0.85 |
ENSMUST00000047899.13
ENSMUST00000164902.8 |
Mettl17
|
methyltransferase like 17 |
| chr14_-_101878106 | 0.81 |
ENSMUST00000100339.9
|
Commd6
|
COMM domain containing 6 |
| chr3_-_95778679 | 0.81 |
ENSMUST00000142437.2
ENSMUST00000067298.5 |
Mrps21
|
mitochondrial ribosomal protein S21 |
| chr11_+_119989736 | 0.81 |
ENSMUST00000106223.4
ENSMUST00000185558.2 |
Ndufaf8
|
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
| chr14_-_101877870 | 0.78 |
ENSMUST00000168587.3
|
Commd6
|
COMM domain containing 6 |
| chr1_+_167135933 | 0.78 |
ENSMUST00000195015.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr11_+_58221569 | 0.76 |
ENSMUST00000073128.7
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr13_+_4241149 | 0.75 |
ENSMUST00000021634.4
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr5_+_147797258 | 0.73 |
ENSMUST00000031654.10
|
Pomp
|
proteasome maturation protein |
| chr17_+_46421908 | 0.72 |
ENSMUST00000024763.10
ENSMUST00000123646.2 |
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr16_+_13721016 | 0.72 |
ENSMUST00000128757.8
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr4_-_118266416 | 0.72 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr5_+_147797425 | 0.70 |
ENSMUST00000201376.4
ENSMUST00000201120.2 |
Pomp
|
proteasome maturation protein |
| chr11_-_96807273 | 0.68 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr15_+_58805605 | 0.67 |
ENSMUST00000022980.5
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr19_-_6134703 | 0.66 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr19_+_3332901 | 0.64 |
ENSMUST00000025745.10
ENSMUST00000025743.7 |
Mrpl21
|
mitochondrial ribosomal protein L21 |
| chr8_-_125589697 | 0.64 |
ENSMUST00000034465.9
|
2810004N23Rik
|
RIKEN cDNA 2810004N23 gene |
| chr10_+_80008768 | 0.63 |
ENSMUST00000041882.7
|
Fam174c
|
family with sequence similarity 174, member C |
| chr14_+_52122439 | 0.62 |
ENSMUST00000167984.2
|
Mettl17
|
methyltransferase like 17 |
| chr10_-_67120959 | 0.59 |
ENSMUST00000159002.2
ENSMUST00000077839.13 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chr5_-_145077048 | 0.59 |
ENSMUST00000031627.9
|
Pdap1
|
PDGFA associated protein 1 |
| chr11_+_58221538 | 0.58 |
ENSMUST00000116376.9
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr4_+_48279794 | 0.58 |
ENSMUST00000030029.10
|
Invs
|
inversin |
| chr8_+_87350672 | 0.58 |
ENSMUST00000034141.18
ENSMUST00000122188.10 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr2_-_168072493 | 0.58 |
ENSMUST00000109193.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr3_-_94794256 | 0.57 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr1_+_58625539 | 0.57 |
ENSMUST00000027193.9
|
Ndufb3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr11_-_96807192 | 0.57 |
ENSMUST00000144731.8
ENSMUST00000127048.8 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr6_-_35516700 | 0.56 |
ENSMUST00000201026.2
ENSMUST00000031866.9 |
Mtpn
|
myotrophin |
| chr2_-_168072295 | 0.56 |
ENSMUST00000154111.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr11_+_106146966 | 0.53 |
ENSMUST00000021049.9
|
Psmc5
|
protease (prosome, macropain) 26S subunit, ATPase 5 |
| chr17_+_35340431 | 0.53 |
ENSMUST00000173380.2
ENSMUST00000173043.4 ENSMUST00000165306.3 |
Gpank1
|
G patch domain and ankyrin repeats 1 |
| chr7_-_126183716 | 0.53 |
ENSMUST00000150311.8
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr7_-_105393519 | 0.52 |
ENSMUST00000141116.2
|
Taf10
|
TATA-box binding protein associated factor 10 |
| chr11_+_115054157 | 0.51 |
ENSMUST00000021077.4
|
Slc9a3r1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
| chrX_-_93256291 | 0.51 |
ENSMUST00000050328.15
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr7_-_67294943 | 0.50 |
ENSMUST00000190276.7
ENSMUST00000032775.12 ENSMUST00000053950.10 ENSMUST00000189836.2 |
Lrrc28
|
leucine rich repeat containing 28 |
| chr14_-_78970160 | 0.50 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr2_-_84545504 | 0.50 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr9_+_20556088 | 0.50 |
ENSMUST00000162303.8
ENSMUST00000161486.8 |
Ubl5
|
ubiquitin-like 5 |
| chr5_-_149559737 | 0.50 |
ENSMUST00000200805.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_-_36283326 | 0.49 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr5_-_139812592 | 0.49 |
ENSMUST00000182602.2
ENSMUST00000031531.14 |
Psmg3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr13_-_4329421 | 0.48 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr9_+_20556147 | 0.48 |
ENSMUST00000161882.8
|
Ubl5
|
ubiquitin-like 5 |
| chr4_+_118477994 | 0.47 |
ENSMUST00000030501.15
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr7_+_79910948 | 0.47 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr19_+_8897732 | 0.47 |
ENSMUST00000096243.7
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr2_+_132528851 | 0.47 |
ENSMUST00000061891.11
|
Shld1
|
shieldin complex subunit 1 |
| chr15_-_58805537 | 0.47 |
ENSMUST00000226931.2
ENSMUST00000228538.3 ENSMUST00000110155.3 |
Tatdn1
|
TatD DNase domain containing 1 |
| chr7_+_65693350 | 0.46 |
ENSMUST00000206065.2
|
Gm45213
|
predicted gene 45213 |
| chr12_-_81579614 | 0.46 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr19_-_44994824 | 0.46 |
ENSMUST00000097715.4
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr4_-_40948196 | 0.46 |
ENSMUST00000030125.5
ENSMUST00000108089.8 ENSMUST00000191273.7 |
Bag1
|
BCL2-associated athanogene 1 |
| chr3_-_65300000 | 0.45 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr11_-_96807233 | 0.45 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr3_+_89960121 | 0.45 |
ENSMUST00000160640.8
ENSMUST00000029552.13 ENSMUST00000162114.8 ENSMUST00000068798.13 |
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr11_-_16458069 | 0.45 |
ENSMUST00000109641.2
|
Sec61g
|
SEC61, gamma subunit |
| chr9_-_57672124 | 0.44 |
ENSMUST00000215158.2
|
Clk3
|
CDC-like kinase 3 |
| chr8_-_22888604 | 0.43 |
ENSMUST00000033871.8
|
Slc25a15
|
solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 |
| chrX_+_106860083 | 0.43 |
ENSMUST00000143975.8
ENSMUST00000144695.8 ENSMUST00000167154.2 |
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr9_-_79667129 | 0.43 |
ENSMUST00000034881.8
|
Cox7a2
|
cytochrome c oxidase subunit 7A2 |
| chrX_-_168103266 | 0.43 |
ENSMUST00000033717.9
ENSMUST00000112115.2 |
Hccs
|
holocytochrome c synthetase |
| chr5_+_145077172 | 0.43 |
ENSMUST00000162594.8
ENSMUST00000162308.8 ENSMUST00000159018.8 ENSMUST00000160075.2 |
Bud31
|
BUD31 homolog |
| chr10_+_79505203 | 0.42 |
ENSMUST00000020552.8
ENSMUST00000239401.2 |
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr6_-_113354826 | 0.42 |
ENSMUST00000032410.14
|
Tada3
|
transcriptional adaptor 3 |
| chr15_-_80449330 | 0.42 |
ENSMUST00000229110.2
|
Enthd1
|
ENTH domain containing 1 |
| chr2_-_132657891 | 0.41 |
ENSMUST00000039554.7
|
Trmt6
|
tRNA methyltransferase 6 |
| chr1_+_4878046 | 0.41 |
ENSMUST00000027036.11
ENSMUST00000150971.8 ENSMUST00000119612.9 ENSMUST00000137887.8 ENSMUST00000115529.8 |
Lypla1
|
lysophospholipase 1 |
| chrX_-_106859842 | 0.41 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr12_+_78908466 | 0.40 |
ENSMUST00000071230.8
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha |
| chr9_+_107440484 | 0.40 |
ENSMUST00000193418.2
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr6_+_83055581 | 0.40 |
ENSMUST00000177177.8
ENSMUST00000176089.2 |
Pcgf1
|
polycomb group ring finger 1 |
| chr11_+_5905693 | 0.39 |
ENSMUST00000002818.9
|
Ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr17_+_47022384 | 0.39 |
ENSMUST00000002840.9
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr9_-_57672375 | 0.39 |
ENSMUST00000215233.2
|
Clk3
|
CDC-like kinase 3 |
| chr19_-_45648751 | 0.39 |
ENSMUST00000046869.11
|
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr10_+_128641398 | 0.39 |
ENSMUST00000217745.2
|
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr11_+_62349238 | 0.37 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr4_+_124696394 | 0.37 |
ENSMUST00000175875.2
|
Mtf1
|
metal response element binding transcription factor 1 |
| chr12_+_104998895 | 0.37 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr8_-_107783282 | 0.37 |
ENSMUST00000034391.4
ENSMUST00000095517.12 |
Cog8
|
component of oligomeric golgi complex 8 |
| chr7_-_4606104 | 0.36 |
ENSMUST00000049113.14
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr19_-_6134903 | 0.36 |
ENSMUST00000160977.8
ENSMUST00000159859.2 ENSMUST00000025707.9 ENSMUST00000160712.8 ENSMUST00000237738.2 |
Zfpl1
|
zinc finger like protein 1 |
| chr11_-_16458484 | 0.36 |
ENSMUST00000109643.8
ENSMUST00000166950.8 ENSMUST00000178855.8 |
Sec61g
|
SEC61, gamma subunit |
| chr9_+_46184362 | 0.36 |
ENSMUST00000156440.8
ENSMUST00000114552.4 |
Zpr1
|
ZPR1 zinc finger |
| chr1_+_155911136 | 0.35 |
ENSMUST00000111757.10
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr15_-_85695855 | 0.35 |
ENSMUST00000134631.8
ENSMUST00000154814.2 ENSMUST00000071876.13 ENSMUST00000150995.8 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr7_+_5083212 | 0.35 |
ENSMUST00000098845.10
ENSMUST00000146317.8 ENSMUST00000153169.2 ENSMUST00000045277.7 |
Epn1
|
epsin 1 |
| chrX_+_143100987 | 0.35 |
ENSMUST00000123710.8
ENSMUST00000149330.8 |
Alg13
|
asparagine-linked glycosylation 13 |
| chr3_-_88332401 | 0.35 |
ENSMUST00000168755.7
ENSMUST00000193433.6 ENSMUST00000195657.6 ENSMUST00000057935.9 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr19_+_8849000 | 0.35 |
ENSMUST00000096255.7
|
Ubxn1
|
UBX domain protein 1 |
| chr12_-_44257109 | 0.35 |
ENSMUST00000015049.5
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr2_+_132658055 | 0.35 |
ENSMUST00000028831.15
ENSMUST00000066559.6 |
Mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr11_-_16458148 | 0.34 |
ENSMUST00000109642.8
|
Sec61g
|
SEC61, gamma subunit |
| chr19_+_8848924 | 0.34 |
ENSMUST00000238036.2
|
Ubxn1
|
UBX domain protein 1 |
| chr19_+_8848876 | 0.34 |
ENSMUST00000166407.9
|
Ubxn1
|
UBX domain protein 1 |
| chr1_+_91178017 | 0.34 |
ENSMUST00000059743.12
ENSMUST00000178627.8 ENSMUST00000171165.8 ENSMUST00000191368.7 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr11_-_50101592 | 0.34 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chrX_-_99669507 | 0.34 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11 |
| chr5_+_124690908 | 0.33 |
ENSMUST00000071057.14
ENSMUST00000111438.2 |
Ddx55
|
DEAD box helicase 55 |
| chr5_-_115487005 | 0.33 |
ENSMUST00000040154.9
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chrX_-_140508177 | 0.33 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr4_+_40948407 | 0.33 |
ENSMUST00000030128.6
|
Chmp5
|
charged multivesicular body protein 5 |
| chr19_-_11833365 | 0.33 |
ENSMUST00000079875.4
|
Olfr1418
|
olfactory receptor 1418 |
| chrX_-_167456828 | 0.32 |
ENSMUST00000033725.16
ENSMUST00000112137.2 |
Msl3
|
MSL complex subunit 3 |
| chr12_-_78908409 | 0.32 |
ENSMUST00000021536.9
|
Atp6v1d
|
ATPase, H+ transporting, lysosomal V1 subunit D |
| chr2_-_130266162 | 0.32 |
ENSMUST00000089581.11
|
Pced1a
|
PC-esterase domain containing 1A |
| chr10_+_128641520 | 0.32 |
ENSMUST00000219508.2
ENSMUST00000026410.2 |
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr19_-_11806388 | 0.32 |
ENSMUST00000061235.3
|
Olfr1417
|
olfactory receptor 1417 |
| chr1_+_4878460 | 0.31 |
ENSMUST00000131119.2
|
Lypla1
|
lysophospholipase 1 |
| chr1_-_179373826 | 0.31 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr6_+_83055321 | 0.31 |
ENSMUST00000165164.9
ENSMUST00000092614.9 |
Pcgf1
|
polycomb group ring finger 1 |
| chr1_+_91178288 | 0.31 |
ENSMUST00000171112.8
ENSMUST00000191533.2 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chrX_+_7750261 | 0.31 |
ENSMUST00000115660.12
|
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr7_-_46569662 | 0.31 |
ENSMUST00000143413.3
ENSMUST00000014546.15 |
Tsg101
|
tumor susceptibility gene 101 |
| chr4_-_118477960 | 0.31 |
ENSMUST00000071972.11
|
Cfap57
|
cilia and flagella associated protein 57 |
| chr5_-_34792361 | 0.31 |
ENSMUST00000202065.2
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr12_-_44256843 | 0.31 |
ENSMUST00000220421.2
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr11_-_95037089 | 0.30 |
ENSMUST00000021241.8
|
Dlx4
|
distal-less homeobox 4 |
| chr15_+_5215180 | 0.30 |
ENSMUST00000081640.12
|
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr9_+_64939695 | 0.30 |
ENSMUST00000034960.14
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr2_-_90735171 | 0.29 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr1_+_74640706 | 0.29 |
ENSMUST00000087186.11
|
Stk36
|
serine/threonine kinase 36 |
| chr9_+_107440445 | 0.29 |
ENSMUST00000010198.5
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr2_+_145776697 | 0.29 |
ENSMUST00000116398.8
ENSMUST00000126415.8 |
Cfap61
|
cilia and flagella associated protein 61 |
| chr2_+_38898065 | 0.29 |
ENSMUST00000112862.7
|
Arpc5l
|
actin related protein 2/3 complex, subunit 5-like |
| chr2_+_145776720 | 0.28 |
ENSMUST00000152515.8
ENSMUST00000138774.8 ENSMUST00000130168.8 ENSMUST00000133433.8 ENSMUST00000118002.2 |
Cfap61
|
cilia and flagella associated protein 61 |
| chr8_+_121854566 | 0.28 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr14_+_47535717 | 0.28 |
ENSMUST00000166743.9
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr4_-_98271469 | 0.28 |
ENSMUST00000143116.2
ENSMUST00000030292.12 ENSMUST00000102793.11 |
Tm2d1
|
TM2 domain containing 1 |
| chr9_-_37259629 | 0.28 |
ENSMUST00000217238.3
|
Ccdc15
|
coiled-coil domain containing 15 |
| chr7_+_81412695 | 0.28 |
ENSMUST00000133034.2
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr7_+_81412673 | 0.27 |
ENSMUST00000042166.11
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr2_+_179899166 | 0.27 |
ENSMUST00000059080.7
|
Rps21
|
ribosomal protein S21 |
| chrX_+_7750483 | 0.27 |
ENSMUST00000115663.10
ENSMUST00000096514.11 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr2_+_160730076 | 0.27 |
ENSMUST00000109457.3
|
Lpin3
|
lipin 3 |
| chr2_-_93841152 | 0.27 |
ENSMUST00000111240.8
|
Alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr9_+_117869543 | 0.27 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr11_-_120515799 | 0.27 |
ENSMUST00000106183.3
ENSMUST00000080202.12 |
Sirt7
|
sirtuin 7 |
| chr18_+_61688329 | 0.26 |
ENSMUST00000165123.8
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr13_+_46822992 | 0.26 |
ENSMUST00000099547.4
|
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr12_-_55126882 | 0.26 |
ENSMUST00000021406.6
|
2700097O09Rik
|
RIKEN cDNA 2700097O09 gene |
| chr7_+_36397426 | 0.26 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr15_-_85695810 | 0.26 |
ENSMUST00000144067.8
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr11_+_94544593 | 0.26 |
ENSMUST00000025278.8
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr11_-_76969230 | 0.25 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1 |
| chr3_-_95125051 | 0.25 |
ENSMUST00000107204.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr5_-_139812162 | 0.25 |
ENSMUST00000182839.2
|
Gm26938
|
predicted gene, 26938 |
| chr2_-_10085299 | 0.25 |
ENSMUST00000114897.9
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr17_-_35340006 | 0.25 |
ENSMUST00000174306.8
ENSMUST00000174024.8 |
Csnk2b
|
casein kinase 2, beta polypeptide |
| chr2_-_127050161 | 0.24 |
ENSMUST00000056146.2
|
1810024B03Rik
|
RIKEN cDNA 1810024B03 gene |
| chr7_+_44499818 | 0.24 |
ENSMUST00000136232.2
ENSMUST00000207223.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr8_-_108315024 | 0.24 |
ENSMUST00000044106.6
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr2_-_93841062 | 0.24 |
ENSMUST00000040005.13
ENSMUST00000126378.8 |
Alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr6_+_124785621 | 0.24 |
ENSMUST00000047760.10
|
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr15_+_9140614 | 0.24 |
ENSMUST00000227556.3
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr6_+_124785834 | 0.24 |
ENSMUST00000143040.8
ENSMUST00000052727.5 ENSMUST00000130160.2 |
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr9_+_57818243 | 0.24 |
ENSMUST00000216925.2
ENSMUST00000163329.2 ENSMUST00000213654.2 ENSMUST00000217132.2 ENSMUST00000216841.2 ENSMUST00000214086.2 |
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr17_-_26063488 | 0.23 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr11_+_88095222 | 0.23 |
ENSMUST00000118784.8
ENSMUST00000139170.8 ENSMUST00000107915.10 ENSMUST00000144070.3 |
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr11_+_88095206 | 0.23 |
ENSMUST00000024486.14
|
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr7_+_112026712 | 0.23 |
ENSMUST00000106643.8
ENSMUST00000033030.14 |
Parva
|
parvin, alpha |
| chr2_-_10085133 | 0.23 |
ENSMUST00000145530.8
ENSMUST00000026887.14 ENSMUST00000114896.8 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr17_-_35340189 | 0.23 |
ENSMUST00000025246.13
ENSMUST00000173114.8 |
Csnk2b
|
casein kinase 2, beta polypeptide |
| chr11_+_87482971 | 0.23 |
ENSMUST00000103179.10
ENSMUST00000092802.12 ENSMUST00000146871.8 |
Mtmr4
|
myotubularin related protein 4 |
| chr4_+_118478357 | 0.23 |
ENSMUST00000147373.2
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr10_-_30476658 | 0.23 |
ENSMUST00000019927.7
|
Trmt11
|
tRNA methyltransferase 11 |
| chr15_+_5215000 | 0.23 |
ENSMUST00000118193.8
ENSMUST00000022751.15 |
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr5_-_110928436 | 0.22 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr9_-_21061196 | 0.22 |
ENSMUST00000215296.2
ENSMUST00000019615.11 |
Cdc37
|
cell division cycle 37 |
| chr15_+_76215488 | 0.22 |
ENSMUST00000172281.8
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr9_+_122576325 | 0.22 |
ENSMUST00000178679.3
|
Topaz1
|
testis and ovary specific PAZ domain containing 1 |
| chr4_+_106479624 | 0.22 |
ENSMUST00000047922.3
|
Ttc22
|
tetratricopeptide repeat domain 22 |
| chr10_+_128641663 | 0.22 |
ENSMUST00000218511.2
|
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr14_-_55909314 | 0.22 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr14_-_55909527 | 0.22 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr17_-_26063391 | 0.22 |
ENSMUST00000176591.8
|
Rhot2
|
ras homolog family member T2 |
| chr6_-_113354668 | 0.22 |
ENSMUST00000193384.2
|
Tada3
|
transcriptional adaptor 3 |
| chr9_+_57818384 | 0.22 |
ENSMUST00000217129.2
|
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr9_+_117869642 | 0.21 |
ENSMUST00000134433.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr2_-_10084866 | 0.21 |
ENSMUST00000130067.2
ENSMUST00000139810.8 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr4_+_107035566 | 0.21 |
ENSMUST00000030361.11
|
Tmem59
|
transmembrane protein 59 |
| chr4_+_107035615 | 0.21 |
ENSMUST00000128123.3
ENSMUST00000106753.3 |
Tmem59
|
transmembrane protein 59 |
| chr19_-_28941264 | 0.21 |
ENSMUST00000161813.2
|
Spata6l
|
spermatogenesis associated 6 like |
| chr2_+_160730019 | 0.21 |
ENSMUST00000109455.9
ENSMUST00000040872.13 |
Lpin3
|
lipin 3 |
| chr7_-_46569617 | 0.20 |
ENSMUST00000210664.2
ENSMUST00000156335.9 |
Tsg101
|
tumor susceptibility gene 101 |
| chr18_+_62681982 | 0.20 |
ENSMUST00000055725.12
ENSMUST00000162365.8 |
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr11_+_87628356 | 0.20 |
ENSMUST00000093955.12
|
Supt4a
|
SPT4A, DSIF elongation factor subunit |
| chr2_-_37333109 | 0.20 |
ENSMUST00000102789.3
ENSMUST00000067043.5 ENSMUST00000112932.2 |
Zbtb26
Zbtb6
|
zinc finger and BTB domain containing 26 zinc finger and BTB domain containing 6 |
| chr5_-_138262178 | 0.20 |
ENSMUST00000048421.14
|
Map11
|
microtubule associated protein 11 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.4 | 3.5 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.2 | 0.7 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 0.7 | GO:0052564 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.2 | 1.7 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.2 | 0.5 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.2 | 1.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.0 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.4 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 0.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.9 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.3 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.5 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.7 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.6 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 1.6 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.3 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.4 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.5 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 1.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.5 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.8 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.3 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.6 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.3 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.4 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.4 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.2 | GO:0000239 | pachytene(GO:0000239) meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.1 | 0.2 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 0.2 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.6 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 2.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.5 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.5 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.9 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.9 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.5 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.6 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.5 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 1.6 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 1.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.4 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.6 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.7 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 2.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 1.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0031428 | dense fibrillar component(GO:0001651) box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.4 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 4.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 1.0 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.2 | 0.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 0.5 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.2 | 0.5 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.1 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 1.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.9 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.6 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 2.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 2.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |