Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Evx2
Z-value: 0.33
Transcription factors associated with Evx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Evx2
|
ENSMUSG00000001815.16 | Evx2 |
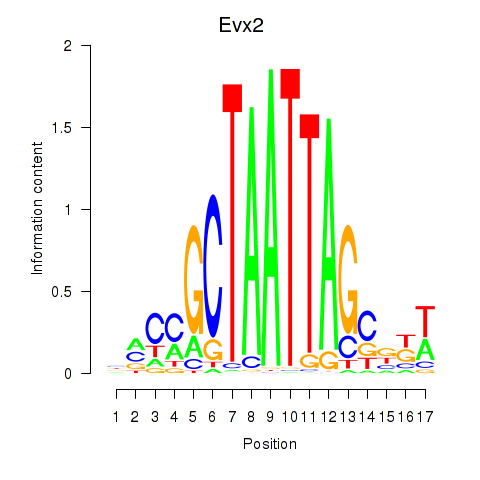
Activity profile of Evx2 motif
Sorted Z-values of Evx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Evx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_154916367 | 1.26 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr1_+_36800874 | 1.03 |
ENSMUST00000027291.7
|
Zap70
|
zeta-chain (TCR) associated protein kinase |
| chr7_-_48493388 | 0.92 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr7_+_51528788 | 0.82 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr6_+_116627635 | 0.59 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chr14_-_64654397 | 0.58 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr11_-_113600346 | 0.54 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr9_-_107556823 | 0.52 |
ENSMUST00000010205.9
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr10_-_128425519 | 0.49 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr2_-_132089667 | 0.42 |
ENSMUST00000110163.8
ENSMUST00000180286.2 ENSMUST00000028816.9 |
Tmem230
|
transmembrane protein 230 |
| chr9_+_47441471 | 0.39 |
ENSMUST00000114548.8
ENSMUST00000152459.8 ENSMUST00000143026.9 ENSMUST00000085909.9 ENSMUST00000114547.8 ENSMUST00000239368.2 ENSMUST00000214542.2 ENSMUST00000034581.4 |
Cadm1
|
cell adhesion molecule 1 |
| chr11_-_113600838 | 0.35 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr1_+_91729175 | 0.31 |
ENSMUST00000007949.4
ENSMUST00000186075.2 |
Twist2
|
twist basic helix-loop-helix transcription factor 2 |
| chr2_+_91087668 | 0.29 |
ENSMUST00000111349.9
ENSMUST00000131711.8 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chrX_+_57075981 | 0.29 |
ENSMUST00000088627.11
|
Zic3
|
zinc finger protein of the cerebellum 3 |
| chrM_+_10167 | 0.28 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr10_+_39488930 | 0.28 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr9_-_50472605 | 0.27 |
ENSMUST00000034568.7
|
Tex12
|
testis expressed 12 |
| chr11_+_116734104 | 0.27 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr7_+_141503411 | 0.27 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr4_+_100336003 | 0.26 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr2_+_91087156 | 0.26 |
ENSMUST00000144394.8
ENSMUST00000028694.12 ENSMUST00000168916.8 ENSMUST00000156919.8 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr9_-_50472620 | 0.26 |
ENSMUST00000217236.2
|
Tex12
|
testis expressed 12 |
| chr14_-_36820304 | 0.23 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chrX_+_57076359 | 0.22 |
ENSMUST00000088631.11
ENSMUST00000088629.4 |
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr16_-_45544960 | 0.22 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr11_-_61157986 | 0.22 |
ENSMUST00000066277.10
ENSMUST00000074127.14 ENSMUST00000108715.3 |
Aldh3a2
|
aldehyde dehydrogenase family 3, subfamily A2 |
| chr10_+_80538115 | 0.21 |
ENSMUST00000218184.2
|
Izumo4
|
IZUMO family member 4 |
| chr18_+_12874390 | 0.21 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr7_-_24423715 | 0.21 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr4_-_42661893 | 0.20 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr19_+_5138562 | 0.20 |
ENSMUST00000238093.2
ENSMUST00000025811.6 ENSMUST00000237025.2 |
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr7_+_141503583 | 0.19 |
ENSMUST00000172652.8
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr2_-_84545504 | 0.18 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr18_+_24737009 | 0.17 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr3_+_63148887 | 0.17 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr1_+_172383499 | 0.16 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr6_+_122603369 | 0.15 |
ENSMUST00000049644.9
|
Dppa3
|
developmental pluripotency-associated 3 |
| chr14_-_59602882 | 0.15 |
ENSMUST00000160425.8
ENSMUST00000095157.11 |
Phf11d
|
PHD finger protein 11D |
| chr14_-_59602859 | 0.15 |
ENSMUST00000161031.2
|
Phf11d
|
PHD finger protein 11D |
| chrM_+_7758 | 0.14 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr3_-_129518723 | 0.12 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr6_-_68968278 | 0.12 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr14_-_44987797 | 0.11 |
ENSMUST00000226900.2
|
Gm8267
|
predicted gene 8267 |
| chr11_-_65160810 | 0.11 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chrX_-_110446022 | 0.11 |
ENSMUST00000156639.2
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr6_-_69394425 | 0.11 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chrM_+_7779 | 0.10 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr16_+_91184661 | 0.10 |
ENSMUST00000139503.2
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr10_-_128885867 | 0.10 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr8_+_114362181 | 0.10 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr17_-_37409147 | 0.10 |
ENSMUST00000216376.2
ENSMUST00000217372.2 |
Olfr91
|
olfactory receptor 91 |
| chr19_-_5610628 | 0.10 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr2_+_167345009 | 0.09 |
ENSMUST00000078050.7
|
Rnf114
|
ring finger protein 114 |
| chr11_+_116547932 | 0.09 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chr6_-_69261303 | 0.08 |
ENSMUST00000103349.2
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr1_-_92423800 | 0.08 |
ENSMUST00000204766.2
ENSMUST00000204009.3 |
Olfr1415
|
olfactory receptor 1415 |
| chr8_+_114362419 | 0.08 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr3_-_15640045 | 0.08 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr17_+_46807637 | 0.08 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr2_-_86208737 | 0.08 |
ENSMUST00000217435.2
|
Olfr1057
|
olfactory receptor 1057 |
| chr6_-_69678271 | 0.07 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr3_-_151541603 | 0.07 |
ENSMUST00000106126.2
|
Ptgfr
|
prostaglandin F receptor |
| chr6_-_69377328 | 0.07 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr2_-_164427367 | 0.07 |
ENSMUST00000109342.2
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr3_-_15491482 | 0.07 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr6_-_124756645 | 0.07 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr17_+_38143840 | 0.07 |
ENSMUST00000213857.2
|
Olfr125
|
olfactory receptor 125 |
| chr7_+_6225277 | 0.07 |
ENSMUST00000072662.12
ENSMUST00000155314.2 |
Zscan5b
|
zinc finger and SCAN domain containing 5B |
| chr1_+_135768409 | 0.07 |
ENSMUST00000189826.7
|
Tnnt2
|
troponin T2, cardiac |
| chr11_-_65160767 | 0.06 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr17_-_36773221 | 0.06 |
ENSMUST00000169950.2
ENSMUST00000057502.14 |
H2-M10.4
|
histocompatibility 2, M region locus 10.4 |
| chr12_+_52746158 | 0.06 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr7_+_6441687 | 0.06 |
ENSMUST00000218906.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr16_+_3648742 | 0.06 |
ENSMUST00000214238.2
ENSMUST00000214590.2 |
Olfr15
|
olfactory receptor 15 |
| chr15_-_9529898 | 0.05 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr6_-_69355456 | 0.05 |
ENSMUST00000196595.2
|
Igkv4-63
|
immunoglobulin kappa variable 4-63 |
| chr7_+_114342929 | 0.05 |
ENSMUST00000161800.2
|
Insc
|
INSC spindle orientation adaptor protein |
| chr11_-_120269517 | 0.05 |
ENSMUST00000026448.10
|
Faap100
|
Fanconi anemia core complex associated protein 100 |
| chr3_+_94744844 | 0.05 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr6_-_69162381 | 0.05 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr2_+_118877610 | 0.04 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr1_-_92412835 | 0.04 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr14_+_73411249 | 0.04 |
ENSMUST00000166875.2
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr6_-_68994064 | 0.04 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr13_+_38388904 | 0.03 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr7_-_86016045 | 0.03 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr19_-_13827773 | 0.03 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr5_-_84565218 | 0.03 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr19_-_13828056 | 0.03 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr17_+_88748139 | 0.03 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr14_+_53521353 | 0.03 |
ENSMUST00000103625.3
|
Trav3n-3
|
T cell receptor alpha variable 3N-3 |
| chr10_+_70010839 | 0.02 |
ENSMUST00000156001.8
ENSMUST00000135607.2 |
Ccdc6
|
coiled-coil domain containing 6 |
| chr7_+_12700030 | 0.02 |
ENSMUST00000210619.2
|
Zfp324
|
zinc finger protein 324 |
| chr6_-_69204417 | 0.02 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr10_+_112151245 | 0.02 |
ENSMUST00000218445.2
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chrX_-_166907286 | 0.02 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr19_+_41921903 | 0.02 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chr11_-_6217718 | 0.01 |
ENSMUST00000004507.11
ENSMUST00000151446.2 |
Ddx56
|
DEAD box helicase 56 |
| chr17_-_37430949 | 0.01 |
ENSMUST00000214994.2
ENSMUST00000216341.2 |
Olfr92
|
olfactory receptor 92 |
| chr2_-_111820618 | 0.01 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr7_+_29515485 | 0.01 |
ENSMUST00000178162.3
ENSMUST00000032796.14 |
Zfp790
|
zinc finger protein 790 |
| chr6_-_68887922 | 0.01 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr2_-_86257093 | 0.01 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr17_+_37148015 | 0.01 |
ENSMUST00000179968.8
ENSMUST00000130367.8 ENSMUST00000053434.15 ENSMUST00000130801.8 ENSMUST00000144182.8 ENSMUST00000123715.8 |
Trim26
|
tripartite motif-containing 26 |
| chr19_+_12364643 | 0.01 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr15_-_102275403 | 0.00 |
ENSMUST00000229464.2
|
Sp7
|
Sp7 transcription factor 7 |
| chr6_-_69020489 | 0.00 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr6_-_69245427 | 0.00 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr17_+_17622934 | 0.00 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr2_-_86109346 | 0.00 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.3 | 0.9 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.2 | 1.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.1 | 0.5 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.5 | GO:0051342 | sensory perception of umami taste(GO:0050917) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.1 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.2 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.2 | GO:2000722 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.2 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |