Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
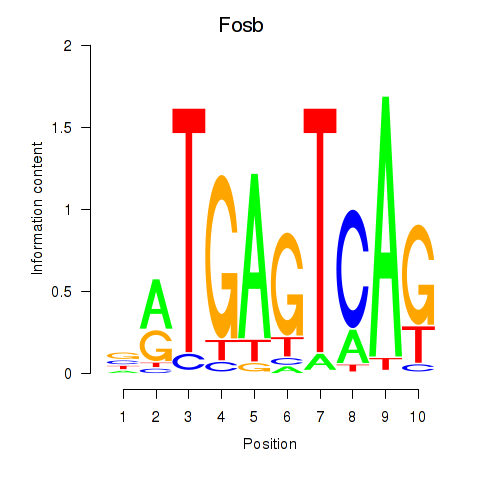
Results for Fosb
Z-value: 0.57
Transcription factors associated with Fosb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosb
|
ENSMUSG00000003545.4 | Fosb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fosb | mm39_v1_chr7_-_19043525_19043627 | -0.10 | 5.8e-01 | Click! |
Activity profile of Fosb motif
Sorted Z-values of Fosb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Fosb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_19922599 | 0.80 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr5_+_31274064 | 0.60 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr5_+_31274046 | 0.56 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr17_-_45903410 | 0.43 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr18_+_12637217 | 0.40 |
ENSMUST00000188815.2
|
Lama3
|
laminin, alpha 3 |
| chr17_-_45903188 | 0.39 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_-_133384449 | 0.37 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr14_+_66872699 | 0.36 |
ENSMUST00000159365.8
ENSMUST00000054661.8 ENSMUST00000225182.2 ENSMUST00000159068.2 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr8_-_110766009 | 0.34 |
ENSMUST00000212934.2
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr11_-_95966407 | 0.33 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr8_-_110765983 | 0.33 |
ENSMUST00000109222.4
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr15_-_75886166 | 0.32 |
ENSMUST00000060807.12
|
Fam83h
|
family with sequence similarity 83, member H |
| chr11_-_83957889 | 0.32 |
ENSMUST00000108101.8
|
Dusp14
|
dual specificity phosphatase 14 |
| chr11_-_100139728 | 0.31 |
ENSMUST00000007280.9
|
Krt16
|
keratin 16 |
| chr11_-_99213769 | 0.30 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr11_-_95966477 | 0.30 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr7_+_126376319 | 0.30 |
ENSMUST00000132643.2
|
Ypel3
|
yippee like 3 |
| chr6_+_129510331 | 0.30 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr19_-_32080496 | 0.29 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr8_+_36956345 | 0.28 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr3_+_102377234 | 0.28 |
ENSMUST00000035952.5
ENSMUST00000198168.5 ENSMUST00000106925.9 |
Ngf
|
nerve growth factor |
| chr9_+_108782664 | 0.27 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr19_+_5927876 | 0.27 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr7_+_139414057 | 0.26 |
ENSMUST00000026548.14
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr13_+_30843937 | 0.26 |
ENSMUST00000091672.13
|
Dusp22
|
dual specificity phosphatase 22 |
| chr17_-_30831576 | 0.26 |
ENSMUST00000235171.2
ENSMUST00000236335.2 ENSMUST00000167624.2 |
Glo1
|
glyoxalase 1 |
| chr1_-_51955126 | 0.26 |
ENSMUST00000046390.14
|
Myo1b
|
myosin IB |
| chr16_+_32219324 | 0.25 |
ENSMUST00000115149.3
|
Tm4sf19
|
transmembrane 4 L six family member 19 |
| chr6_-_87327885 | 0.25 |
ENSMUST00000032129.3
|
Gkn1
|
gastrokine 1 |
| chr19_-_38113249 | 0.25 |
ENSMUST00000112335.4
|
Rbp4
|
retinol binding protein 4, plasma |
| chr11_+_51541728 | 0.25 |
ENSMUST00000117859.8
ENSMUST00000064493.6 |
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr3_-_59102517 | 0.25 |
ENSMUST00000200095.2
|
Gpr87
|
G protein-coupled receptor 87 |
| chr1_-_74932266 | 0.25 |
ENSMUST00000006721.3
|
Cryba2
|
crystallin, beta A2 |
| chr9_+_108782646 | 0.23 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr19_+_5927821 | 0.23 |
ENSMUST00000145200.8
ENSMUST00000025732.14 ENSMUST00000125114.8 ENSMUST00000155697.8 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr2_+_84891281 | 0.23 |
ENSMUST00000238769.2
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr7_-_4781140 | 0.22 |
ENSMUST00000094892.12
|
Il11
|
interleukin 11 |
| chr17_+_48080113 | 0.22 |
ENSMUST00000160373.8
ENSMUST00000159641.8 |
Tfeb
|
transcription factor EB |
| chr9_+_44151962 | 0.22 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr5_-_24597009 | 0.22 |
ENSMUST00000059401.7
|
Atg9b
|
autophagy related 9B |
| chr9_+_7445822 | 0.22 |
ENSMUST00000034497.8
|
Mmp3
|
matrix metallopeptidase 3 |
| chr1_-_10790120 | 0.22 |
ENSMUST00000035577.7
|
Cpa6
|
carboxypeptidase A6 |
| chr3_-_92481033 | 0.21 |
ENSMUST00000053107.6
|
Ivl
|
involucrin |
| chr1_-_75240551 | 0.21 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr9_+_7272514 | 0.21 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr11_+_4207557 | 0.21 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr12_+_111132908 | 0.20 |
ENSMUST00000139162.8
ENSMUST00000060274.7 |
Traf3
|
TNF receptor-associated factor 3 |
| chr1_+_165596961 | 0.20 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr10_-_5872386 | 0.20 |
ENSMUST00000131996.8
ENSMUST00000064225.14 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr7_+_140659038 | 0.19 |
ENSMUST00000159375.8
|
Pkp3
|
plakophilin 3 |
| chr6_+_53264255 | 0.19 |
ENSMUST00000203528.3
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr17_+_36152559 | 0.19 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr19_-_38113056 | 0.19 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr7_-_127545896 | 0.19 |
ENSMUST00000118755.8
ENSMUST00000094026.10 |
Prss36
|
protease, serine 36 |
| chr10_-_85847697 | 0.18 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr7_+_127400016 | 0.18 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr7_-_4974167 | 0.18 |
ENSMUST00000133272.2
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr11_-_100288566 | 0.18 |
ENSMUST00000001592.15
ENSMUST00000107403.2 |
Jup
|
junction plakoglobin |
| chr7_-_80051455 | 0.18 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr7_-_141926079 | 0.18 |
ENSMUST00000059223.15
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr11_-_51541610 | 0.16 |
ENSMUST00000142721.2
ENSMUST00000156835.8 ENSMUST00000001080.16 |
N4bp3
|
NEDD4 binding protein 3 |
| chr10_-_5872341 | 0.16 |
ENSMUST00000117676.8
ENSMUST00000019909.8 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr16_+_17379749 | 0.16 |
ENSMUST00000171002.10
ENSMUST00000023441.11 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr7_+_30487322 | 0.16 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr8_+_4276827 | 0.16 |
ENSMUST00000053035.7
|
Lrrc8e
|
leucine rich repeat containing 8 family, member E |
| chr6_-_124746445 | 0.16 |
ENSMUST00000156033.2
|
Eno2
|
enolase 2, gamma neuronal |
| chr18_+_82929037 | 0.16 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr16_+_20367327 | 0.16 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr4_+_109263820 | 0.16 |
ENSMUST00000124209.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr4_+_42158092 | 0.15 |
ENSMUST00000098122.3
|
Gm13306
|
predicted gene 13306 |
| chr15_-_74650673 | 0.15 |
ENSMUST00000186014.2
ENSMUST00000191407.2 |
Ly6g6g
|
lymphocyte antigen 6 complex, locus G6G |
| chr13_+_93440572 | 0.15 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr7_+_26135039 | 0.15 |
ENSMUST00000119386.8
ENSMUST00000146907.3 |
Nlrp4a
|
NLR family, pyrin domain containing 4A |
| chr13_+_15638466 | 0.15 |
ENSMUST00000110510.4
|
Gli3
|
GLI-Kruppel family member GLI3 |
| chr8_+_108020132 | 0.15 |
ENSMUST00000151114.8
ENSMUST00000125721.8 ENSMUST00000075922.11 |
Nfat5
|
nuclear factor of activated T cells 5 |
| chr2_-_90900628 | 0.15 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr15_+_102412157 | 0.15 |
ENSMUST00000096145.5
|
Gm10337
|
predicted gene 10337 |
| chr11_-_69439934 | 0.14 |
ENSMUST00000108659.2
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr19_+_5497575 | 0.14 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr19_-_38113696 | 0.14 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr3_+_55369384 | 0.14 |
ENSMUST00000200352.2
|
Dclk1
|
doublecortin-like kinase 1 |
| chr19_-_4189603 | 0.14 |
ENSMUST00000025761.8
|
Cabp4
|
calcium binding protein 4 |
| chr9_+_114807546 | 0.14 |
ENSMUST00000183104.8
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr12_+_16944896 | 0.13 |
ENSMUST00000020904.8
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2 |
| chr14_-_52257452 | 0.13 |
ENSMUST00000228162.2
|
Zfp219
|
zinc finger protein 219 |
| chr8_+_108020092 | 0.13 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr4_-_140393185 | 0.13 |
ENSMUST00000069623.12
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr3_-_79439181 | 0.13 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr9_+_78522783 | 0.13 |
ENSMUST00000093812.5
|
Cd109
|
CD109 antigen |
| chr6_-_124746468 | 0.13 |
ENSMUST00000204896.3
|
Eno2
|
enolase 2, gamma neuronal |
| chr3_+_55369149 | 0.12 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr15_-_74599860 | 0.12 |
ENSMUST00000023261.4
ENSMUST00000190433.2 |
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr7_+_75879603 | 0.12 |
ENSMUST00000156166.8
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr6_-_124746510 | 0.12 |
ENSMUST00000149652.2
ENSMUST00000112476.8 ENSMUST00000004378.15 |
Eno2
|
enolase 2, gamma neuronal |
| chr10_-_43880353 | 0.12 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr11_+_80319424 | 0.12 |
ENSMUST00000173938.8
ENSMUST00000017572.14 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr9_+_104443876 | 0.12 |
ENSMUST00000157006.8
|
Cpne4
|
copine IV |
| chr7_-_144305705 | 0.11 |
ENSMUST00000155175.8
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr17_-_43053057 | 0.11 |
ENSMUST00000239223.2
ENSMUST00000113614.3 |
Adgrf2
|
adhesion G protein-coupled receptor F2 |
| chr10_-_18619439 | 0.11 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chr18_+_44204457 | 0.11 |
ENSMUST00000068473.4
ENSMUST00000236634.2 |
Spink6
|
serine peptidase inhibitor, Kazal type 6 |
| chr12_+_71062733 | 0.11 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr8_-_117809188 | 0.11 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr7_+_126376353 | 0.11 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr3_+_55369288 | 0.10 |
ENSMUST00000198412.5
ENSMUST00000199169.5 ENSMUST00000199702.5 ENSMUST00000198437.5 |
Dclk1
|
doublecortin-like kinase 1 |
| chr7_+_6048160 | 0.10 |
ENSMUST00000037728.13
ENSMUST00000121583.2 |
Nlrp4c
|
NLR family, pyrin domain containing 4C |
| chr7_+_127160751 | 0.10 |
ENSMUST00000190278.3
|
Tmem265
|
transmembrane protein 265 |
| chr2_-_18053595 | 0.10 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr6_-_135095237 | 0.10 |
ENSMUST00000032327.14
ENSMUST00000111922.2 |
Gprc5d
|
G protein-coupled receptor, family C, group 5, member D |
| chr1_-_13061333 | 0.10 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr2_-_151474391 | 0.10 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chr7_-_119461027 | 0.10 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr2_+_118493713 | 0.09 |
ENSMUST00000099557.10
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr10_+_87926932 | 0.08 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr1_+_34116308 | 0.08 |
ENSMUST00000239001.2
|
Dst
|
dystonin |
| chr2_+_103242027 | 0.08 |
ENSMUST00000239273.2
ENSMUST00000164172.8 |
Elf5
|
E74-like factor 5 |
| chr9_+_7347369 | 0.08 |
ENSMUST00000005950.12
ENSMUST00000065079.6 |
Mmp12
|
matrix metallopeptidase 12 |
| chr18_+_82928782 | 0.08 |
ENSMUST00000235793.2
|
Zfp516
|
zinc finger protein 516 |
| chr18_+_42186757 | 0.08 |
ENSMUST00000074679.4
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr10_-_18619658 | 0.08 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr10_-_63926044 | 0.08 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr11_-_120358239 | 0.08 |
ENSMUST00000076921.7
|
Arl16
|
ADP-ribosylation factor-like 16 |
| chr18_+_42186713 | 0.08 |
ENSMUST00000072008.11
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr17_-_36008863 | 0.07 |
ENSMUST00000146472.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr7_-_126073994 | 0.07 |
ENSMUST00000205733.2
ENSMUST00000205889.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr13_+_93441307 | 0.07 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr3_-_122828592 | 0.07 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr2_-_29142965 | 0.07 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chrX_-_74621828 | 0.07 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr18_-_31450095 | 0.07 |
ENSMUST00000139924.2
ENSMUST00000153060.8 |
Rit2
|
Ras-like without CAAX 2 |
| chr1_-_36574801 | 0.06 |
ENSMUST00000054665.10
ENSMUST00000194853.3 |
Ankrd23
|
ankyrin repeat domain 23 |
| chr7_+_30463175 | 0.06 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr19_+_4189786 | 0.06 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr15_+_101371353 | 0.05 |
ENSMUST00000088049.5
|
Krt86
|
keratin 86 |
| chr2_+_51928017 | 0.05 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr5_+_93004330 | 0.05 |
ENSMUST00000225438.2
|
Shroom3
|
shroom family member 3 |
| chr15_-_74544409 | 0.05 |
ENSMUST00000023268.14
ENSMUST00000110009.4 |
Arc
|
activity regulated cytoskeletal-associated protein |
| chr17_-_24863956 | 0.04 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr4_-_132459762 | 0.04 |
ENSMUST00000045550.5
|
Xkr8
|
X-linked Kx blood group related 8 |
| chr11_+_77692116 | 0.04 |
ENSMUST00000100794.10
ENSMUST00000151373.4 ENSMUST00000130305.9 ENSMUST00000172303.10 ENSMUST00000092884.11 ENSMUST00000164334.8 |
Myo18a
|
myosin XVIIIA |
| chr17_+_78815493 | 0.04 |
ENSMUST00000024880.11
ENSMUST00000232859.2 |
Vit
|
vitrin |
| chr13_-_113800172 | 0.03 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chr5_-_122640255 | 0.03 |
ENSMUST00000031423.10
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr10_+_115979787 | 0.03 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr7_+_30475819 | 0.03 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr14_-_55995912 | 0.03 |
ENSMUST00000001497.9
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr2_+_151947444 | 0.03 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr3_+_64884839 | 0.03 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr17_-_24863907 | 0.02 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr7_+_126376099 | 0.02 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr9_+_122717536 | 0.02 |
ENSMUST00000063980.8
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr8_-_70426910 | 0.01 |
ENSMUST00000116463.4
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr16_-_4698148 | 0.00 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr5_-_122639840 | 0.00 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr5_+_32293145 | 0.00 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 0.4 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.6 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.2 | GO:0060367 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.0 | 0.3 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.2 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 1.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 1.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.6 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |