Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
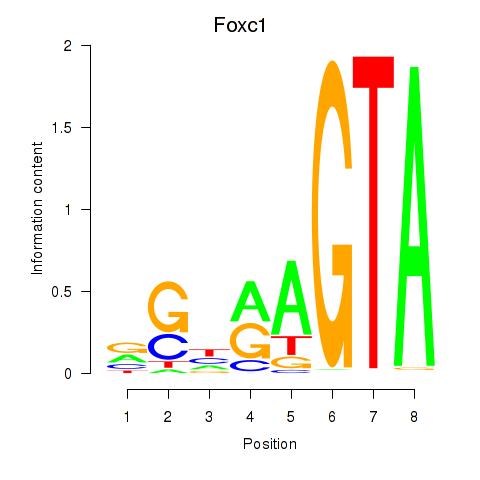
Results for Foxc1
Z-value: 0.73
Transcription factors associated with Foxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc1
|
ENSMUSG00000050295.5 | Foxc1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc1 | mm39_v1_chr13_+_31990604_31990633 | -0.14 | 4.0e-01 | Click! |
Activity profile of Foxc1 motif
Sorted Z-values of Foxc1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_105775224 | 3.47 |
ENSMUST00000093222.13
ENSMUST00000093223.5 |
Ces3a
|
carboxylesterase 3A |
| chr9_-_103099262 | 3.24 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr11_+_69945157 | 3.14 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr19_+_40078132 | 2.26 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr1_-_172722589 | 2.11 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr6_-_55152002 | 2.06 |
ENSMUST00000003569.6
|
Inmt
|
indolethylamine N-methyltransferase |
| chr14_-_31362835 | 1.98 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr14_-_31362909 | 1.94 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr11_+_108286114 | 1.58 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr1_+_88093726 | 1.51 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr2_+_58645189 | 1.43 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr2_+_58644922 | 1.21 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr8_+_105810380 | 1.15 |
ENSMUST00000093221.13
ENSMUST00000074403.13 |
Ces3b
|
carboxylesterase 3B |
| chr3_+_94284739 | 1.10 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr5_+_146016064 | 1.08 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr8_-_25066313 | 0.99 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr14_+_31363004 | 0.92 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chr7_+_141996067 | 0.89 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr17_-_31348576 | 0.89 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr10_-_89369432 | 0.89 |
ENSMUST00000105297.2
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr1_+_131890679 | 0.87 |
ENSMUST00000191034.2
ENSMUST00000177943.8 |
Gm29103
Slc45a3
|
predicted gene 29103 solute carrier family 45, member 3 |
| chr3_+_94284812 | 0.84 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr2_-_10135449 | 0.78 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chrX_+_100492684 | 0.77 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr3_-_122828592 | 0.67 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr12_+_104067346 | 0.65 |
ENSMUST00000021495.4
|
Serpina5
|
serine (or cysteine) peptidase inhibitor, clade A, member 5 |
| chr11_+_100973391 | 0.64 |
ENSMUST00000001806.10
ENSMUST00000107308.4 |
Coasy
|
Coenzyme A synthase |
| chr7_+_119360141 | 0.63 |
ENSMUST00000106528.8
ENSMUST00000106527.8 ENSMUST00000063770.10 ENSMUST00000106529.8 |
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr15_+_10177709 | 0.58 |
ENSMUST00000124470.8
|
Prlr
|
prolactin receptor |
| chr2_+_30127692 | 0.56 |
ENSMUST00000113654.8
ENSMUST00000095078.3 |
Lrrc8a
|
leucine rich repeat containing 8A VRAC subunit A |
| chr2_-_165242307 | 0.53 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr15_-_100497863 | 0.53 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr11_-_3881789 | 0.52 |
ENSMUST00000109992.8
ENSMUST00000109988.2 |
Tcn2
|
transcobalamin 2 |
| chr1_+_135768595 | 0.50 |
ENSMUST00000112087.9
ENSMUST00000178854.8 ENSMUST00000027671.12 ENSMUST00000179863.8 ENSMUST00000112085.9 ENSMUST00000112086.3 |
Tnnt2
|
troponin T2, cardiac |
| chr12_-_113386312 | 0.49 |
ENSMUST00000177715.8
ENSMUST00000103426.3 |
Ighm
|
immunoglobulin heavy constant mu |
| chr8_+_118428643 | 0.48 |
ENSMUST00000034304.9
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr1_+_45350698 | 0.47 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr9_-_44920899 | 0.46 |
ENSMUST00000102832.3
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr2_+_172212642 | 0.44 |
ENSMUST00000116375.2
|
Cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr8_-_106660470 | 0.44 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr6_+_113449237 | 0.44 |
ENSMUST00000204447.3
|
Il17rc
|
interleukin 17 receptor C |
| chr17_+_57318469 | 0.42 |
ENSMUST00000210548.2
|
Gm17949
|
predicted gene, 17949 |
| chr16_-_94171340 | 0.40 |
ENSMUST00000138514.2
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr9_+_108447077 | 0.38 |
ENSMUST00000019183.14
|
Dalrd3
|
DALR anticodon binding domain containing 3 |
| chr15_-_78290038 | 0.38 |
ENSMUST00000058659.9
|
Tst
|
thiosulfate sulfurtransferase, mitochondrial |
| chr13_-_30039613 | 0.37 |
ENSMUST00000091674.12
ENSMUST00000006353.14 |
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr19_-_40982576 | 0.36 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr7_+_114344920 | 0.35 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr1_+_135980508 | 0.35 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr7_+_30193047 | 0.35 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr17_+_34457868 | 0.34 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr4_+_97665992 | 0.34 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr1_+_135980488 | 0.33 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr7_+_18962301 | 0.31 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chr4_+_106924181 | 0.31 |
ENSMUST00000106758.8
ENSMUST00000145324.8 ENSMUST00000106760.8 |
Cyb5rl
|
cytochrome b5 reductase-like |
| chr18_-_3281089 | 0.30 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr11_-_78441584 | 0.29 |
ENSMUST00000103242.5
|
Tmem97
|
transmembrane protein 97 |
| chr9_-_51240201 | 0.29 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr8_-_84299540 | 0.28 |
ENSMUST00000212990.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr8_-_69541852 | 0.28 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr2_-_155356716 | 0.28 |
ENSMUST00000029131.11
|
Ggt7
|
gamma-glutamyltransferase 7 |
| chr11_-_84058292 | 0.27 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr15_-_57897702 | 0.27 |
ENSMUST00000100655.5
ENSMUST00000185553.2 |
9130401M01Rik
|
RIKEN cDNA 9130401M01 gene |
| chr17_+_46565116 | 0.26 |
ENSMUST00000095262.6
|
Lrrc73
|
leucine rich repeat containing 73 |
| chrX_+_156481906 | 0.26 |
ENSMUST00000136141.2
ENSMUST00000190091.7 |
Smpx
|
small muscle protein, X-linked |
| chr2_+_30697838 | 0.26 |
ENSMUST00000041830.10
ENSMUST00000152374.8 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chrX_-_94521712 | 0.25 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr2_-_181007099 | 0.24 |
ENSMUST00000108808.8
ENSMUST00000170190.8 ENSMUST00000127988.8 |
Arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr4_+_100336003 | 0.24 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr7_-_5128936 | 0.23 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr8_-_84769170 | 0.23 |
ENSMUST00000005601.9
|
Il27ra
|
interleukin 27 receptor, alpha |
| chr16_+_17149235 | 0.22 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr17_+_37356872 | 0.22 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr9_-_89586960 | 0.22 |
ENSMUST00000058488.9
|
Tmed3
|
transmembrane p24 trafficking protein 3 |
| chr2_+_181007177 | 0.22 |
ENSMUST00000108807.9
|
Zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr19_-_11301919 | 0.21 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr7_+_48895879 | 0.21 |
ENSMUST00000064395.13
|
Nav2
|
neuron navigator 2 |
| chr10_+_128540049 | 0.21 |
ENSMUST00000217836.2
|
Pmel
|
premelanosome protein |
| chr4_+_117407517 | 0.19 |
ENSMUST00000037127.15
|
Eri3
|
exoribonuclease 3 |
| chr15_-_102165884 | 0.18 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma |
| chr15_+_76784110 | 0.18 |
ENSMUST00000068407.6
ENSMUST00000109793.3 |
Commd5
|
COMM domain containing 5 |
| chr3_-_96104886 | 0.17 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chr7_+_18962252 | 0.17 |
ENSMUST00000063976.9
|
Opa3
|
optic atrophy 3 |
| chr12_-_73093953 | 0.17 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr9_+_120762466 | 0.17 |
ENSMUST00000007130.15
ENSMUST00000178812.9 |
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr3_-_57202301 | 0.16 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr17_+_37356854 | 0.16 |
ENSMUST00000025338.16
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr9_-_106768601 | 0.16 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr4_+_43267165 | 0.16 |
ENSMUST00000107942.9
ENSMUST00000102953.4 |
Atp8b5
|
ATPase, class I, type 8B, member 5 |
| chr14_+_53980561 | 0.16 |
ENSMUST00000103667.6
|
Trav16
|
T cell receptor alpha variable 16 |
| chr18_+_37948443 | 0.16 |
ENSMUST00000195239.2
|
Pcdhgc4
|
protocadherin gamma subfamily C, 4 |
| chr18_+_65022035 | 0.15 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr6_-_67512768 | 0.15 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr1_-_131025562 | 0.15 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr2_+_129854256 | 0.15 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr10_-_116732813 | 0.14 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr4_+_97665843 | 0.14 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr18_-_3280999 | 0.14 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr2_+_172212577 | 0.13 |
ENSMUST00000151511.8
|
Cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr12_-_85421467 | 0.13 |
ENSMUST00000040766.9
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr10_+_14581325 | 0.13 |
ENSMUST00000191238.7
ENSMUST00000190114.2 |
Nmbr
|
neuromedin B receptor |
| chr5_-_24732200 | 0.12 |
ENSMUST00000088311.6
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr6_-_68746087 | 0.12 |
ENSMUST00000103333.4
|
Igkv4-91
|
immunoglobulin kappa chain variable 4-91 |
| chr6_+_41118120 | 0.12 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr1_+_135768409 | 0.12 |
ENSMUST00000189826.7
|
Tnnt2
|
troponin T2, cardiac |
| chr12_-_101943134 | 0.11 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr13_-_18556626 | 0.11 |
ENSMUST00000139064.10
ENSMUST00000175703.9 |
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr9_-_71499628 | 0.11 |
ENSMUST00000093823.8
|
Myzap
|
myocardial zonula adherens protein |
| chr2_+_70392351 | 0.11 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr15_+_102412157 | 0.10 |
ENSMUST00000096145.5
|
Gm10337
|
predicted gene 10337 |
| chr17_+_35056219 | 0.10 |
ENSMUST00000173995.2
ENSMUST00000174092.8 |
Dxo
|
decapping exoribonuclease |
| chr17_-_24908874 | 0.10 |
ENSMUST00000007236.5
|
Syngr3
|
synaptogyrin 3 |
| chr9_-_71499594 | 0.10 |
ENSMUST00000166112.2
|
Myzap
|
myocardial zonula adherens protein |
| chr9_-_96513529 | 0.10 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr16_+_37360245 | 0.09 |
ENSMUST00000023524.13
|
Rabl3
|
RAB, member RAS oncogene family-like 3 |
| chr5_-_33591320 | 0.09 |
ENSMUST00000173348.2
|
Nkx1-1
|
NK1 homeobox 1 |
| chrX_-_163033364 | 0.09 |
ENSMUST00000112263.2
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr2_-_79738734 | 0.09 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr4_-_94817025 | 0.09 |
ENSMUST00000030309.6
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr2_-_79738773 | 0.09 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr16_-_48592372 | 0.08 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr2_+_172392678 | 0.08 |
ENSMUST00000099058.10
|
Tfap2c
|
transcription factor AP-2, gamma |
| chr16_-_19703014 | 0.08 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr9_+_123635556 | 0.08 |
ENSMUST00000216072.2
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr1_-_87322443 | 0.08 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr1_+_36753607 | 0.08 |
ENSMUST00000208994.2
|
4933424G06Rik
|
RIKEN cDNA 4933424G06 gene |
| chr16_-_59452883 | 0.08 |
ENSMUST00000118438.8
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr17_+_56266146 | 0.07 |
ENSMUST00000086869.6
|
Yju2
|
YJU2 splicing factor |
| chr16_-_48592319 | 0.07 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr6_-_93889483 | 0.07 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr4_-_49521036 | 0.06 |
ENSMUST00000057829.4
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr16_+_43067641 | 0.06 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr11_+_67131403 | 0.06 |
ENSMUST00000170942.2
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr13_+_42834039 | 0.06 |
ENSMUST00000128646.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr17_-_52117894 | 0.06 |
ENSMUST00000176669.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr3_-_89905547 | 0.06 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr9_-_71499547 | 0.05 |
ENSMUST00000169573.8
|
Myzap
|
myocardial zonula adherens protein |
| chr6_-_30680502 | 0.05 |
ENSMUST00000133373.8
|
Cep41
|
centrosomal protein 41 |
| chrX_+_119199956 | 0.05 |
ENSMUST00000113364.10
ENSMUST00000050239.16 ENSMUST00000113358.10 |
Pcdh11x
|
protocadherin 11 X-linked |
| chr17_+_56266099 | 0.05 |
ENSMUST00000233661.2
|
Yju2
|
YJU2 splicing factor |
| chr3_+_95067759 | 0.05 |
ENSMUST00000131742.8
ENSMUST00000090823.8 ENSMUST00000090821.10 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr11_+_104983022 | 0.05 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr18_-_37771772 | 0.05 |
ENSMUST00000058635.5
|
Slc25a2
|
solute carrier family 25 (mitochondrial carrier, ornithine transporter) member 2 |
| chr18_+_65021974 | 0.05 |
ENSMUST00000225261.3
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr6_+_29402868 | 0.05 |
ENSMUST00000154619.5
|
Ccdc136
|
coiled-coil domain containing 136 |
| chr2_-_104679838 | 0.04 |
ENSMUST00000126824.2
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr5_-_66309244 | 0.04 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr6_-_88604404 | 0.04 |
ENSMUST00000120933.5
|
Kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr16_-_43709968 | 0.04 |
ENSMUST00000023387.14
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr14_+_78141679 | 0.04 |
ENSMUST00000022591.16
ENSMUST00000169978.2 ENSMUST00000227903.2 |
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr2_-_76045696 | 0.04 |
ENSMUST00000144892.2
|
Pde11a
|
phosphodiesterase 11A |
| chr3_-_37180093 | 0.04 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr8_-_78337297 | 0.04 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr6_+_17281184 | 0.04 |
ENSMUST00000000058.7
|
Cav2
|
caveolin 2 |
| chr17_-_90395568 | 0.04 |
ENSMUST00000173222.2
|
Nrxn1
|
neurexin I |
| chr8_+_84379298 | 0.04 |
ENSMUST00000019577.10
ENSMUST00000211985.2 ENSMUST00000212463.2 |
Gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr2_+_49956441 | 0.04 |
ENSMUST00000112712.10
ENSMUST00000128451.8 ENSMUST00000053208.14 |
Lypd6
|
LY6/PLAUR domain containing 6 |
| chr3_-_57202546 | 0.03 |
ENSMUST00000196506.2
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr3_-_92514799 | 0.03 |
ENSMUST00000195278.2
|
2210017I01Rik
|
RIKEN cDNA 2210017I01 gene |
| chr6_-_52190299 | 0.03 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308 |
| chr9_-_104214899 | 0.03 |
ENSMUST00000112590.3
|
Acpp
|
acid phosphatase, prostate |
| chr5_+_88635834 | 0.03 |
ENSMUST00000199104.5
ENSMUST00000031222.9 |
Enam
|
enamelin |
| chrX_+_65696608 | 0.03 |
ENSMUST00000036043.5
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr9_-_110305705 | 0.03 |
ENSMUST00000198164.5
ENSMUST00000068025.13 |
Klhl18
|
kelch-like 18 |
| chr17_+_3447465 | 0.02 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr6_-_83513184 | 0.02 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr15_+_6451721 | 0.02 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr1_+_75213044 | 0.02 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr6_-_120271520 | 0.02 |
ENSMUST00000057283.8
ENSMUST00000212457.2 |
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr14_+_63235512 | 0.02 |
ENSMUST00000100492.5
|
Defb47
|
defensin beta 47 |
| chr16_-_96971905 | 0.02 |
ENSMUST00000056102.9
|
Dscam
|
DS cell adhesion molecule |
| chr7_+_30487322 | 0.02 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr6_-_88022172 | 0.02 |
ENSMUST00000203674.3
ENSMUST00000204126.2 ENSMUST00000113596.8 ENSMUST00000113600.10 |
Rab7
|
RAB7, member RAS oncogene family |
| chr14_+_53167246 | 0.02 |
ENSMUST00000177703.3
|
Trav12d-3
|
T cell receptor alpha variable 12D-3 |
| chr13_-_54914366 | 0.02 |
ENSMUST00000036825.14
|
Sncb
|
synuclein, beta |
| chr14_+_53370136 | 0.02 |
ENSMUST00000181793.3
|
Trav6n-6
|
T cell receptor alpha variable 6N-6 |
| chr8_-_65146079 | 0.02 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr14_-_48900192 | 0.02 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr9_-_95288775 | 0.02 |
ENSMUST00000036267.8
|
Chst2
|
carbohydrate sulfotransferase 2 |
| chr11_+_63022328 | 0.02 |
ENSMUST00000018361.10
|
Pmp22
|
peripheral myelin protein 22 |
| chr1_-_4430481 | 0.01 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr2_-_103315483 | 0.01 |
ENSMUST00000028610.10
|
Cat
|
catalase |
| chr3_-_89905927 | 0.01 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr4_-_94817056 | 0.01 |
ENSMUST00000107097.9
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr14_-_50425655 | 0.01 |
ENSMUST00000205837.3
|
Olfr730
|
olfactory receptor 730 |
| chr14_-_36690636 | 0.01 |
ENSMUST00000067700.13
|
Ccser2
|
coiled-coil serine rich 2 |
| chr5_+_114313940 | 0.01 |
ENSMUST00000146841.2
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr14_+_53059379 | 0.01 |
ENSMUST00000197754.2
|
Trav6d-6
|
T cell receptor alpha variable 6D-6 |
| chr1_+_163822438 | 0.01 |
ENSMUST00000045694.14
ENSMUST00000111490.2 |
Mettl18
|
methyltransferase like 18 |
| chr15_+_89218601 | 0.01 |
ENSMUST00000023282.9
|
Miox
|
myo-inositol oxygenase |
| chr6_-_77956499 | 0.01 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr4_-_58009118 | 0.01 |
ENSMUST00000102897.11
ENSMUST00000239406.2 |
Txndc8
|
thioredoxin domain containing 8 |
| chr10_+_115405891 | 0.01 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr5_-_30401416 | 0.01 |
ENSMUST00000125367.4
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chrX_-_74460137 | 0.00 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr8_+_120301974 | 0.00 |
ENSMUST00000093100.3
|
Dnaaf1
|
dynein, axonemal assembly factor 1 |
| chr18_+_37884653 | 0.00 |
ENSMUST00000194928.2
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr1_+_170060318 | 0.00 |
ENSMUST00000162752.2
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr11_-_96720309 | 0.00 |
ENSMUST00000167149.8
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.6 | 3.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 3.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.3 | 2.8 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.2 | 0.9 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 1.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 2.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 0.5 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.2 | 0.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.5 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.4 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 1.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.0 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.1 | 0.2 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.3 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.2 | GO:1904793 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 0.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.2 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.5 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.2 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.9 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 2.9 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.0 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.2 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 2.1 | GO:0009308 | amine metabolic process(GO:0009308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.2 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 2.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.7 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.5 | 3.2 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.4 | 2.6 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.3 | 1.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.3 | 3.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.3 | 1.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 2.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 1.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 2.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.9 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 1.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.2 | 0.6 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 0.9 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 3.2 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 4.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.2 | GO:1990226 | alpha-catenin binding(GO:0045294) histone methyltransferase binding(GO:1990226) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |