Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxf1
Z-value: 1.02
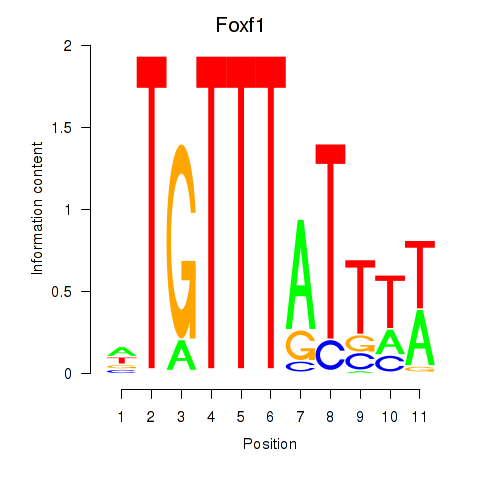
Transcription factors associated with Foxf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxf1
|
ENSMUSG00000042812.6 | Foxf1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf1 | mm39_v1_chr8_+_121811091_121811125 | -0.18 | 3.0e-01 | Click! |
Activity profile of Foxf1 motif
Sorted Z-values of Foxf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_12894716 | 5.80 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr15_+_4756684 | 4.33 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr9_-_48516447 | 4.23 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr15_+_4756657 | 4.17 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
| chr2_+_67948057 | 4.12 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr1_-_130589321 | 4.08 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr1_-_130589349 | 4.02 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr6_+_41279199 | 3.47 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr17_-_31363245 | 3.40 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr8_-_93956143 | 3.13 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr1_-_139487951 | 2.96 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr19_+_30210320 | 2.44 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr6_+_30541581 | 2.43 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr18_+_51250748 | 2.28 |
ENSMUST00000116639.4
|
Prr16
|
proline rich 16 |
| chr3_-_148696155 | 2.23 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr17_+_25097199 | 2.19 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr19_-_58443593 | 2.07 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr3_-_20329823 | 2.04 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr18_-_39051695 | 2.02 |
ENSMUST00000040647.11
|
Fgf1
|
fibroblast growth factor 1 |
| chr3_-_10400710 | 1.95 |
ENSMUST00000078748.4
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr19_-_58443830 | 1.92 |
ENSMUST00000026076.14
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr7_-_100306160 | 1.90 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr6_-_59001455 | 1.86 |
ENSMUST00000089860.12
|
Fam13a
|
family with sequence similarity 13, member A |
| chr8_+_46111703 | 1.65 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_+_75088220 | 1.63 |
ENSMUST00000185307.7
ENSMUST00000229521.2 |
Ly6g2
|
lymphocyte antigen 6 complex, locus G2 |
| chr5_-_87054796 | 1.62 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr15_+_75088445 | 1.55 |
ENSMUST00000055719.8
|
Ly6g2
|
lymphocyte antigen 6 complex, locus G2 |
| chr6_+_129510331 | 1.47 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr4_-_57916283 | 1.46 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr14_-_45715308 | 1.44 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr1_+_67162176 | 1.40 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr9_+_53212871 | 1.40 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chrX_+_141011173 | 1.38 |
ENSMUST00000112914.8
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr6_+_129510145 | 1.34 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chrM_+_9870 | 1.34 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr2_+_71884943 | 1.32 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr5_-_98178834 | 1.31 |
ENSMUST00000199088.2
|
Antxr2
|
anthrax toxin receptor 2 |
| chr6_+_129510117 | 1.31 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr9_+_114560235 | 1.30 |
ENSMUST00000035007.10
|
Cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr2_+_67935015 | 1.25 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chrX_+_141010919 | 1.25 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr13_+_24054241 | 1.24 |
ENSMUST00000110413.8
|
Slc17a1
|
solute carrier family 17 (sodium phosphate), member 1 |
| chr4_-_6275629 | 1.23 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr9_-_71070506 | 1.22 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr19_-_58444336 | 1.20 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr11_-_86884507 | 1.19 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr8_-_26275182 | 1.19 |
ENSMUST00000038498.10
|
Bag4
|
BCL2-associated athanogene 4 |
| chr19_+_44980565 | 1.18 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr17_+_37269468 | 1.17 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr1_+_88030951 | 1.14 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr5_+_90708962 | 1.13 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr12_+_59178072 | 1.13 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr3_+_20039775 | 1.12 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr15_+_9071331 | 1.12 |
ENSMUST00000190591.10
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr15_+_3300249 | 1.10 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr6_-_59001325 | 1.10 |
ENSMUST00000173193.2
|
Fam13a
|
family with sequence similarity 13, member A |
| chr3_+_122277372 | 1.09 |
ENSMUST00000197073.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr18_-_3281089 | 1.08 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr19_-_34855242 | 1.07 |
ENSMUST00000238065.2
|
Pank1
|
pantothenate kinase 1 |
| chrM_+_10167 | 1.01 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr4_+_102427247 | 1.01 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr4_+_104623505 | 1.01 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr17_+_37269513 | 0.99 |
ENSMUST00000173814.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr12_+_59178258 | 0.93 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr2_-_91025492 | 0.93 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chrX_-_55643429 | 0.93 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr17_+_29077385 | 0.93 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr5_+_88731366 | 0.91 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr5_+_88731386 | 0.91 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr1_+_34275665 | 0.90 |
ENSMUST00000194192.3
|
Dst
|
dystonin |
| chr17_-_56312555 | 0.89 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr13_-_30039613 | 0.87 |
ENSMUST00000091674.12
ENSMUST00000006353.14 |
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr5_+_102629240 | 0.86 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_+_173579285 | 0.85 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr3_-_137837117 | 0.84 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr18_-_39062201 | 0.81 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr15_+_9071655 | 0.81 |
ENSMUST00000227682.3
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr12_-_108241597 | 0.77 |
ENSMUST00000222310.2
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr5_-_98178811 | 0.76 |
ENSMUST00000031281.14
|
Antxr2
|
anthrax toxin receptor 2 |
| chr11_-_86698484 | 0.76 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr15_+_92495007 | 0.73 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr2_-_51862941 | 0.73 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr16_+_10363254 | 0.73 |
ENSMUST00000115827.8
ENSMUST00000150894.2 ENSMUST00000038145.13 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr7_-_65020955 | 0.72 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr9_-_64160899 | 0.70 |
ENSMUST00000005066.9
|
Map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr4_-_96479793 | 0.70 |
ENSMUST00000055693.9
|
Cyp2j9
|
cytochrome P450, family 2, subfamily j, polypeptide 9 |
| chr1_+_171052623 | 0.70 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr6_-_83633064 | 0.70 |
ENSMUST00000014686.3
|
Clec4f
|
C-type lectin domain family 4, member f |
| chr17_+_45817750 | 0.69 |
ENSMUST00000024733.9
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr15_-_96929086 | 0.69 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr2_-_90309509 | 0.68 |
ENSMUST00000111495.9
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr3_+_85878376 | 0.68 |
ENSMUST00000238443.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr15_-_5137951 | 0.68 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr8_-_84059048 | 0.66 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr17_+_25059079 | 0.66 |
ENSMUST00000164251.8
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr15_-_5137975 | 0.64 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr7_-_65020655 | 0.63 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr2_-_51039112 | 0.62 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr18_-_39062514 | 0.61 |
ENSMUST00000235922.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr15_+_9071761 | 0.61 |
ENSMUST00000189437.8
ENSMUST00000190874.8 |
Nadk2
|
NAD kinase 2, mitochondrial |
| chr19_+_43770619 | 0.60 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr6_-_124410452 | 0.60 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr11_+_99770013 | 0.58 |
ENSMUST00000078442.4
|
Gm11567
|
predicted gene 11567 |
| chr14_-_68819544 | 0.58 |
ENSMUST00000022641.9
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr2_-_134396268 | 0.58 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr8_+_46111778 | 0.57 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr17_+_43671314 | 0.55 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr18_+_32064358 | 0.54 |
ENSMUST00000025254.9
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr16_+_10363222 | 0.54 |
ENSMUST00000155633.8
ENSMUST00000066345.15 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr6_-_136852792 | 0.54 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr7_-_101714251 | 0.53 |
ENSMUST00000130074.2
ENSMUST00000131104.3 ENSMUST00000096639.12 |
Rnf121
|
ring finger protein 121 |
| chr15_+_54274151 | 0.53 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr15_-_37734579 | 0.53 |
ENSMUST00000145909.9
ENSMUST00000153775.9 |
Gm49397
Ncald
|
predicted gene, 49397 neurocalcin delta |
| chr5_+_102629365 | 0.51 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr7_-_48494959 | 0.50 |
ENSMUST00000208050.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr2_-_32314017 | 0.50 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr9_+_44951075 | 0.50 |
ENSMUST00000217097.2
|
Mpzl2
|
myelin protein zero-like 2 |
| chr6_-_52160816 | 0.50 |
ENSMUST00000134831.2
|
Hoxa3
|
homeobox A3 |
| chr1_+_177272215 | 0.49 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr3_+_7431717 | 0.49 |
ENSMUST00000192468.2
ENSMUST00000028999.12 |
Pkia
|
protein kinase inhibitor, alpha |
| chr2_-_51863203 | 0.49 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr12_-_70278188 | 0.48 |
ENSMUST00000161083.2
|
Pygl
|
liver glycogen phosphorylase |
| chr14_+_120513106 | 0.47 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr5_-_66211842 | 0.47 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr12_+_55126999 | 0.47 |
ENSMUST00000220578.2
ENSMUST00000221655.2 |
Srp54a
|
signal recognition particle 54A |
| chr10_-_128759817 | 0.47 |
ENSMUST00000131271.2
|
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr1_-_170978157 | 0.45 |
ENSMUST00000155798.2
ENSMUST00000081560.5 ENSMUST00000111336.10 |
Sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr5_-_90788460 | 0.44 |
ENSMUST00000202704.4
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr18_+_70058533 | 0.42 |
ENSMUST00000043929.11
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr5_-_88823049 | 0.42 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr9_-_44714263 | 0.42 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr1_+_75119472 | 0.40 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr2_+_55327110 | 0.40 |
ENSMUST00000112633.3
ENSMUST00000112632.2 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr1_+_153300874 | 0.40 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr8_-_64659004 | 0.40 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr2_-_144369261 | 0.40 |
ENSMUST00000163701.2
ENSMUST00000081982.12 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr12_+_55201889 | 0.39 |
ENSMUST00000110708.4
|
Srp54b
|
signal recognition particle 54B |
| chr5_-_90788323 | 0.39 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_-_4847673 | 0.39 |
ENSMUST00000066041.12
ENSMUST00000119433.4 |
Shisa7
|
shisa family member 7 |
| chr11_-_46280336 | 0.38 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr19_-_6886965 | 0.38 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr16_-_85347305 | 0.38 |
ENSMUST00000175700.8
ENSMUST00000114174.3 |
Cyyr1
|
cysteine and tyrosine-rich protein 1 |
| chr18_+_70058613 | 0.37 |
ENSMUST00000080050.6
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr15_+_4404965 | 0.37 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr13_-_96678987 | 0.36 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chrX_+_106132840 | 0.35 |
ENSMUST00000118666.8
ENSMUST00000053375.4 |
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr2_+_48704121 | 0.34 |
ENSMUST00000063886.4
|
Acvr2a
|
activin receptor IIA |
| chr7_-_34354924 | 0.33 |
ENSMUST00000032709.3
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr10_-_41455203 | 0.33 |
ENSMUST00000095227.10
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr9_+_122717536 | 0.33 |
ENSMUST00000063980.8
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr2_-_79738734 | 0.33 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_-_101176147 | 0.32 |
ENSMUST00000239140.2
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr2_-_101451383 | 0.32 |
ENSMUST00000090513.11
|
Iftap
|
intraflagellar transport associated protein |
| chr9_-_105398346 | 0.32 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr1_-_43235914 | 0.32 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2 |
| chr13_-_59930059 | 0.32 |
ENSMUST00000225581.2
|
Gm49354
|
predicted gene, 49354 |
| chr6_-_143892814 | 0.31 |
ENSMUST00000124233.7
ENSMUST00000144289.4 ENSMUST00000111748.8 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr1_+_37338964 | 0.31 |
ENSMUST00000027287.11
ENSMUST00000140264.8 |
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I |
| chr4_+_57821050 | 0.31 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr11_-_54751738 | 0.31 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chr17_-_43003135 | 0.31 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr12_+_104180795 | 0.30 |
ENSMUST00000121337.8
ENSMUST00000167049.8 ENSMUST00000101080.2 |
Serpina3f
|
serine (or cysteine) peptidase inhibitor, clade A, member 3F |
| chr16_+_43184191 | 0.30 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_+_75119419 | 0.30 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr17_+_37535982 | 0.29 |
ENSMUST00000078209.4
|
Olfr96
|
olfactory receptor 96 |
| chr15_-_3333003 | 0.29 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr19_-_6887361 | 0.29 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chrX_+_100419965 | 0.29 |
ENSMUST00000119080.8
|
Gjb1
|
gap junction protein, beta 1 |
| chr10_-_12745109 | 0.28 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chrX_-_107877909 | 0.28 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr10_+_25308466 | 0.28 |
ENSMUST00000219224.2
ENSMUST00000219166.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr11_-_46280281 | 0.28 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr3_+_107137924 | 0.28 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr8_+_107329983 | 0.27 |
ENSMUST00000000312.12
ENSMUST00000167688.2 |
Cdh1
|
cadherin 1 |
| chr8_+_106785434 | 0.27 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr11_-_65636651 | 0.27 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr11_-_46280298 | 0.27 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr16_+_43582584 | 0.26 |
ENSMUST00000023390.5
|
Drd3
|
dopamine receptor D3 |
| chr2_-_79738773 | 0.26 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr9_-_100388857 | 0.26 |
ENSMUST00000112874.4
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chr1_+_179788675 | 0.26 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_87365859 | 0.26 |
ENSMUST00000032127.6
|
Gkn3
|
gastrokine 3 |
| chr1_+_58841808 | 0.26 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr19_-_20931566 | 0.25 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr9_-_119548388 | 0.25 |
ENSMUST00000213392.2
|
Scn10a
|
sodium channel, voltage-gated, type X, alpha |
| chr9_+_62249730 | 0.24 |
ENSMUST00000156461.2
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr13_+_83672708 | 0.24 |
ENSMUST00000199105.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr10_+_129072073 | 0.24 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chr14_+_54423447 | 0.24 |
ENSMUST00000103709.2
|
Traj32
|
T cell receptor alpha joining 32 |
| chr17_+_35327255 | 0.24 |
ENSMUST00000037849.3
|
Ly6g5c
|
lymphocyte antigen 6 complex, locus G5C |
| chr11_+_87938128 | 0.24 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr17_+_33387421 | 0.24 |
ENSMUST00000214891.2
ENSMUST00000216633.2 ENSMUST00000214888.2 |
Olfr55
|
olfactory receptor 55 |
| chr10_+_70276604 | 0.23 |
ENSMUST00000173042.9
ENSMUST00000062883.7 |
Fam13c
|
family with sequence similarity 13, member C |
| chr6_+_29361408 | 0.23 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr18_+_46730765 | 0.23 |
ENSMUST00000238168.2
ENSMUST00000078079.11 ENSMUST00000168382.2 ENSMUST00000235849.2 ENSMUST00000235973.2 ENSMUST00000235455.2 ENSMUST00000237478.2 |
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr10_-_61814852 | 0.23 |
ENSMUST00000105453.8
ENSMUST00000105452.9 ENSMUST00000105454.3 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr17_+_37274714 | 0.23 |
ENSMUST00000209623.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr18_+_67266784 | 0.23 |
ENSMUST00000236918.2
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr16_-_95260104 | 0.22 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr19_+_8719033 | 0.22 |
ENSMUST00000176314.8
ENSMUST00000073430.14 ENSMUST00000175901.8 |
Stx5a
|
syntaxin 5A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.0 | 5.8 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.6 | 5.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 3.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.4 | 3.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 1.3 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.4 | 1.2 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.4 | 1.2 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.4 | 1.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.3 | 3.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 1.2 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.3 | 2.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 1.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 4.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.2 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 | 0.9 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.9 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.2 | 0.8 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.2 | 1.4 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 2.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.7 | GO:0046340 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 0.5 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.2 | 0.6 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 1.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 1.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.6 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.1 | 0.5 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.6 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.3 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.5 | GO:0021615 | specification of organ position(GO:0010159) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.3 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 2.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.7 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.2 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.1 | 0.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 0.9 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.7 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 1.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 8.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.2 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) neuroblast migration(GO:0097402) |
| 0.1 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 1.6 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.6 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.3 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.8 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.7 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 2.1 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.7 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.0 | 0.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.5 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.5 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.4 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 2.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 1.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0021941 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0072107 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 2.9 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:1905203 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.0 | 1.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 4.2 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 2.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 2.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 1.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 1.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 2.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 5.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.5 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 2.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.3 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.1 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 6.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 9.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 5.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.3 | 5.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.5 | 1.4 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.4 | 3.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.3 | 0.9 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 1.2 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.2 | 1.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 3.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 4.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 0.8 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.2 | 1.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.2 | 3.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 3.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 0.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 4.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.9 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 1.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 2.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 1.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 2.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 2.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 2.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 4.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.5 | GO:0034236 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 3.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 3.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.0 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.2 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 5.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 3.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 11.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 3.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 3.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 4.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 4.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |