Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
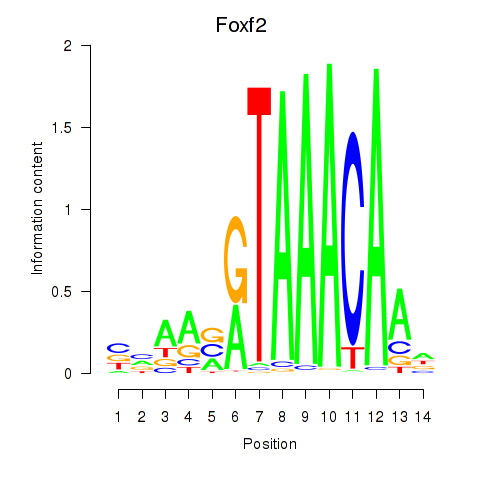
Results for Foxf2
Z-value: 0.56
Transcription factors associated with Foxf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxf2
|
ENSMUSG00000038402.3 | Foxf2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf2 | mm39_v1_chr13_+_31809774_31809799 | 0.07 | 6.8e-01 | Click! |
Activity profile of Foxf2 motif
Sorted Z-values of Foxf2 motif
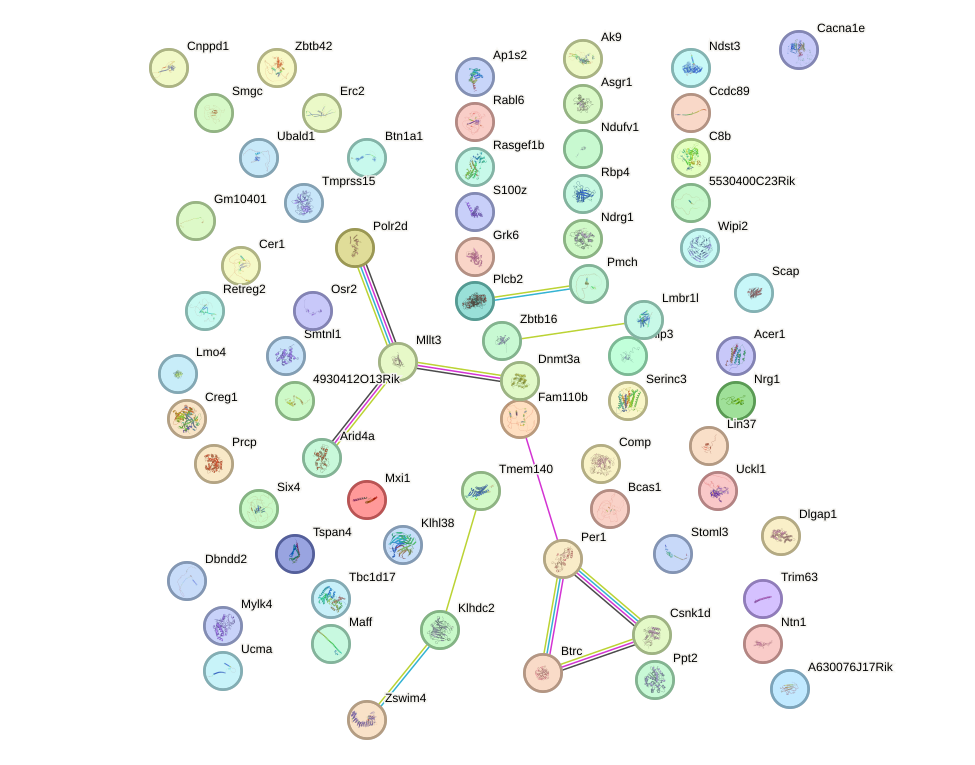
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_66841465 | 1.74 |
ENSMUST00000170903.8
ENSMUST00000166420.8 ENSMUST00000005256.14 ENSMUST00000164070.2 |
Ndrg1
|
N-myc downstream regulated gene 1 |
| chr8_+_70826191 | 1.47 |
ENSMUST00000003659.9
|
Comp
|
cartilage oligomeric matrix protein |
| chr15_+_79231720 | 1.44 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr15_+_79232137 | 1.40 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr17_+_29309942 | 1.12 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr17_+_47816042 | 1.02 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr19_+_45433899 | 0.97 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr17_+_47816074 | 0.94 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr17_+_47815968 | 0.90 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr13_+_55593116 | 0.87 |
ENSMUST00000001115.16
ENSMUST00000224995.2 ENSMUST00000225925.2 ENSMUST00000099482.5 ENSMUST00000224118.2 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr15_+_79232591 | 0.82 |
ENSMUST00000229285.2
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr17_+_47816137 | 0.75 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr12_+_112645237 | 0.72 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr2_-_118558852 | 0.71 |
ENSMUST00000102524.8
|
Plcb2
|
phospholipase C, beta 2 |
| chr2_-_118558825 | 0.70 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
| chr16_-_4698148 | 0.70 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr4_+_104623505 | 0.69 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr1_-_75119277 | 0.65 |
ENSMUST00000168720.8
ENSMUST00000041213.12 ENSMUST00000189809.2 |
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr15_-_98816012 | 0.60 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr7_-_44498305 | 0.52 |
ENSMUST00000207293.2
ENSMUST00000207532.2 |
Tbc1d17
|
TBC1 domain family, member 17 |
| chr2_+_164328375 | 0.50 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr10_+_115653152 | 0.50 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr7_+_29991101 | 0.48 |
ENSMUST00000150892.2
ENSMUST00000126216.2 ENSMUST00000014065.16 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr4_-_87724533 | 0.47 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr11_+_69945157 | 0.46 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr3_-_143908111 | 0.46 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr3_-_143908060 | 0.42 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr5_-_115236354 | 0.41 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr8_+_94537910 | 0.41 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr2_-_181220698 | 0.40 |
ENSMUST00000136875.2
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr12_+_69343450 | 0.39 |
ENSMUST00000021362.5
|
Klhdc2
|
kelch domain containing 2 |
| chr7_-_30259253 | 0.37 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr2_-_25498459 | 0.35 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr8_-_84963653 | 0.35 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr11_+_68986043 | 0.34 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr7_+_44498415 | 0.33 |
ENSMUST00000107885.8
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr8_+_112262729 | 0.33 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr12_+_71063431 | 0.32 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr4_+_5724305 | 0.31 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr18_+_52958382 | 0.31 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr11_-_54846873 | 0.31 |
ENSMUST00000155316.2
ENSMUST00000108889.10 ENSMUST00000126703.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr7_+_44498640 | 0.31 |
ENSMUST00000054343.15
ENSMUST00000142880.3 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr3_-_123483772 | 0.30 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr2_-_163486998 | 0.30 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr2_+_164328763 | 0.30 |
ENSMUST00000109349.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr7_-_141925947 | 0.29 |
ENSMUST00000084412.6
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr12_-_111946560 | 0.29 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
| chr7_+_44499005 | 0.28 |
ENSMUST00000150335.2
ENSMUST00000107882.8 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr15_-_50746202 | 0.27 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr15_+_78209920 | 0.27 |
ENSMUST00000230264.3
|
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr15_+_78210190 | 0.27 |
ENSMUST00000229034.2
ENSMUST00000096355.4 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr15_-_58187556 | 0.26 |
ENSMUST00000022985.2
|
Klhl38
|
kelch-like 38 |
| chr5_+_107656810 | 0.26 |
ENSMUST00000160160.6
|
Gm42669
|
predicted gene 42669 |
| chr7_+_141056305 | 0.24 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr1_+_165596961 | 0.23 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chrX_+_162694397 | 0.23 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr4_+_101365144 | 0.23 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr19_+_53317844 | 0.23 |
ENSMUST00000111737.3
ENSMUST00000025998.15 ENSMUST00000237837.2 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr7_+_90075762 | 0.22 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr8_-_33374282 | 0.21 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chr1_+_75119419 | 0.21 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr10_+_87926932 | 0.21 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr17_-_34846323 | 0.21 |
ENSMUST00000168709.3
ENSMUST00000064953.15 ENSMUST00000170345.8 ENSMUST00000171121.9 ENSMUST00000168391.9 ENSMUST00000169067.9 |
Gm20460
Ppt2
|
predicted gene 20460 palmitoyl-protein thioesterase 2 |
| chr8_-_33374825 | 0.20 |
ENSMUST00000238791.2
|
Nrg1
|
neuregulin 1 |
| chr3_-_37286714 | 0.20 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr2_-_86109346 | 0.20 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr19_-_4062656 | 0.20 |
ENSMUST00000134479.8
ENSMUST00000128787.8 ENSMUST00000237862.2 ENSMUST00000236203.2 ENSMUST00000133474.8 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr17_-_34846122 | 0.20 |
ENSMUST00000171376.8
ENSMUST00000169287.2 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr6_+_15185202 | 0.19 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr9_+_110162470 | 0.19 |
ENSMUST00000198761.5
ENSMUST00000197630.3 |
Scap
|
SREBF chaperone |
| chr6_+_15184630 | 0.19 |
ENSMUST00000115470.3
|
Foxp2
|
forkhead box P2 |
| chr15_+_91722524 | 0.19 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr19_-_4062738 | 0.18 |
ENSMUST00000136921.2
ENSMUST00000042497.14 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr4_+_134042423 | 0.18 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr17_+_47815998 | 0.18 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3 |
| chr14_+_27598021 | 0.17 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr17_+_70829050 | 0.17 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr15_+_78210242 | 0.17 |
ENSMUST00000229678.2
ENSMUST00000231888.2 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr2_+_67004178 | 0.16 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr12_-_25147139 | 0.16 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr19_-_11301919 | 0.16 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr4_-_82803384 | 0.16 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr13_-_23650045 | 0.16 |
ENSMUST00000041674.14
ENSMUST00000110434.2 |
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr4_+_101365052 | 0.15 |
ENSMUST00000038207.12
|
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr16_-_78887971 | 0.15 |
ENSMUST00000023566.11
ENSMUST00000060402.6 |
Tmprss15
|
transmembrane protease, serine 15 |
| chr12_+_3857077 | 0.15 |
ENSMUST00000174817.8
|
Dnmt3a
|
DNA methyltransferase 3A |
| chr15_+_35296237 | 0.14 |
ENSMUST00000022952.6
|
Osr2
|
odd-skipped related 2 |
| chr4_-_87724512 | 0.14 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr17_+_70829144 | 0.14 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
| chr13_-_95615229 | 0.14 |
ENSMUST00000022186.5
|
S100z
|
S100 calcium binding protein, zeta |
| chr3_-_133250889 | 0.14 |
ENSMUST00000197118.5
|
Tet2
|
tet methylcytosine dioxygenase 2 |
| chr6_-_57821483 | 0.13 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr9_-_48747232 | 0.13 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr11_+_85243970 | 0.13 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chr13_-_32967937 | 0.12 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr1_+_83022653 | 0.12 |
ENSMUST00000222567.2
|
Gm47969
|
predicted gene, 47969 |
| chr7_+_92524495 | 0.12 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr5_-_99400698 | 0.12 |
ENSMUST00000031276.15
ENSMUST00000168092.8 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr6_+_34840151 | 0.12 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr16_+_5703134 | 0.11 |
ENSMUST00000230658.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr19_-_38113696 | 0.11 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr2_-_84652890 | 0.11 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr3_+_53396120 | 0.11 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr6_+_34840057 | 0.11 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr3_+_107137924 | 0.10 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr14_+_50618620 | 0.10 |
ENSMUST00000215263.2
ENSMUST00000213402.2 ENSMUST00000213755.2 ENSMUST00000215227.2 |
Olfr736
|
olfactory receptor 736 |
| chr5_+_142615292 | 0.10 |
ENSMUST00000036872.16
ENSMUST00000110778.2 |
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr11_-_68277799 | 0.10 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chr18_+_37827413 | 0.10 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr12_-_73160181 | 0.10 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr1_+_34044940 | 0.10 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr1_+_75119472 | 0.09 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr7_+_29991366 | 0.09 |
ENSMUST00000144508.2
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr5_-_6926523 | 0.09 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr11_-_120881697 | 0.09 |
ENSMUST00000154483.8
|
Csnk1d
|
casein kinase 1, delta |
| chr1_-_154692678 | 0.08 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr2_+_9887427 | 0.08 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr2_-_170248421 | 0.08 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr10_+_41179966 | 0.08 |
ENSMUST00000173494.4
|
Ak9
|
adenylate kinase 9 |
| chr18_+_66006119 | 0.08 |
ENSMUST00000025395.10
|
Grp
|
gastrin releasing peptide |
| chr5_-_103359117 | 0.07 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr10_-_128016135 | 0.07 |
ENSMUST00000238843.2
ENSMUST00000099139.9 |
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr16_-_22258320 | 0.07 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr9_-_44714263 | 0.07 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr1_-_39760150 | 0.07 |
ENSMUST00000151913.3
|
Rfx8
|
regulatory factor X 8 |
| chrX_+_93278203 | 0.07 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15 |
| chr2_+_143757193 | 0.07 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr8_+_59364789 | 0.06 |
ENSMUST00000062978.7
|
BC030500
|
cDNA sequence BC030500 |
| chr18_+_66005891 | 0.06 |
ENSMUST00000173985.10
|
Grp
|
gastrin releasing peptide |
| chr14_+_58310143 | 0.06 |
ENSMUST00000022545.14
|
Fgf9
|
fibroblast growth factor 9 |
| chr12_-_112477536 | 0.06 |
ENSMUST00000066791.7
|
Tmem179
|
transmembrane protein 179 |
| chr14_+_65612788 | 0.05 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr14_+_111912529 | 0.04 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr3_-_144511566 | 0.04 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr8_-_18791557 | 0.04 |
ENSMUST00000033846.7
|
Angpt2
|
angiopoietin 2 |
| chr9_-_99599312 | 0.04 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr12_+_3857001 | 0.04 |
ENSMUST00000020991.15
ENSMUST00000172509.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr13_+_44994167 | 0.04 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr10_+_59942274 | 0.04 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr16_-_22258469 | 0.03 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr1_-_82748964 | 0.03 |
ENSMUST00000223838.2
|
Gm47791
|
predicted gene, 47791 |
| chr3_-_36107661 | 0.03 |
ENSMUST00000166644.8
ENSMUST00000200469.5 |
Ccdc144b
|
coiled-coil domain containing 144B |
| chr3_-_92922976 | 0.03 |
ENSMUST00000107301.2
ENSMUST00000029521.5 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr8_-_37420293 | 0.02 |
ENSMUST00000179501.2
|
Dlc1
|
deleted in liver cancer 1 |
| chr6_-_39702127 | 0.02 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr3_+_54063459 | 0.02 |
ENSMUST00000029311.11
ENSMUST00000200048.5 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr4_+_155875629 | 0.02 |
ENSMUST00000105593.2
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr6_+_146934082 | 0.02 |
ENSMUST00000036194.6
|
Rep15
|
RAB15 effector protein |
| chr7_+_108209994 | 0.02 |
ENSMUST00000209296.3
|
Olfr506
|
olfactory receptor 506 |
| chr10_-_37014859 | 0.02 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr7_+_45433306 | 0.02 |
ENSMUST00000072580.12
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr16_-_34083549 | 0.01 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr18_-_47466378 | 0.01 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr18_-_88945571 | 0.01 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr9_-_58026090 | 0.01 |
ENSMUST00000098681.4
ENSMUST00000098682.10 ENSMUST00000215944.2 |
Ccdc33
|
coiled-coil domain containing 33 |
| chr15_-_66432938 | 0.01 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr2_+_112114906 | 0.00 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 1.7 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 | 3.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.4 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 1.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 1.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.6 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.4 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 1.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.9 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 3.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.6 | GO:0007379 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.9 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.1 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 1.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 3.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 0.9 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.6 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 3.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 4.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 3.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |