Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxl1
Z-value: 0.62
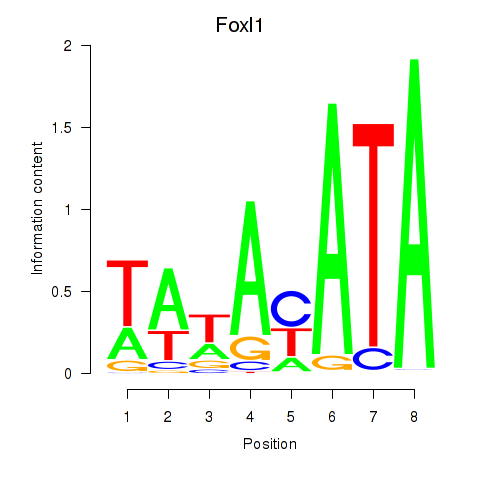
Transcription factors associated with Foxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxl1
|
ENSMUSG00000097084.2 | Foxl1 |
|
Foxl1
|
ENSMUSG00000097084.2 | Foxl1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxl1 | mm39_v1_chr8_+_121854566_121854679 | 0.67 | 6.6e-06 | Click! |
Activity profile of Foxl1 motif
Sorted Z-values of Foxl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41121979 | 4.89 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr11_+_31822211 | 2.03 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_+_11191726 | 1.19 |
ENSMUST00000029866.16
ENSMUST00000108324.4 |
Ccne2
|
cyclin E2 |
| chr9_+_71123061 | 0.99 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr13_+_23922783 | 0.80 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr13_+_23947641 | 0.70 |
ENSMUST00000055770.4
|
H1f1
|
H1.1 linker histone, cluster member |
| chr4_-_120874376 | 0.68 |
ENSMUST00000043200.8
|
Smap2
|
small ArfGAP 2 |
| chr7_-_119078472 | 0.66 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr4_+_11191354 | 0.52 |
ENSMUST00000170901.8
|
Ccne2
|
cyclin E2 |
| chr10_-_127190280 | 0.49 |
ENSMUST00000059718.6
|
Inhbe
|
inhibin beta-E |
| chr3_-_122828592 | 0.47 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr2_+_18059982 | 0.39 |
ENSMUST00000028076.15
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr18_-_15284476 | 0.35 |
ENSMUST00000025992.7
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr8_+_93553901 | 0.32 |
ENSMUST00000034187.9
|
Mmp2
|
matrix metallopeptidase 2 |
| chr6_-_102441628 | 0.32 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr7_-_119078330 | 0.27 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chr13_-_23806530 | 0.27 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr10_-_102326286 | 0.26 |
ENSMUST00000020040.5
|
Nts
|
neurotensin |
| chrX_+_132751729 | 0.26 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr8_+_15107646 | 0.23 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr10_+_87926932 | 0.21 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr10_+_56253418 | 0.20 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr18_+_37864045 | 0.19 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr6_+_106095726 | 0.18 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr11_+_54517164 | 0.17 |
ENSMUST00000239168.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr15_-_11905697 | 0.16 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr16_+_44913974 | 0.16 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr3_-_146475974 | 0.14 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr18_+_37637317 | 0.12 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr1_+_66426127 | 0.12 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr6_-_14755249 | 0.08 |
ENSMUST00000045096.6
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr16_-_22258469 | 0.07 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr1_+_179788037 | 0.05 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_-_169938060 | 0.04 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr5_-_87847268 | 0.04 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr9_-_101076198 | 0.02 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr3_-_82957104 | 0.02 |
ENSMUST00000048246.5
|
Fgb
|
fibrinogen beta chain |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.9 | GO:0072218 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.2 | 2.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.0 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.2 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.3 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 1.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 2.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 2.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |