Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
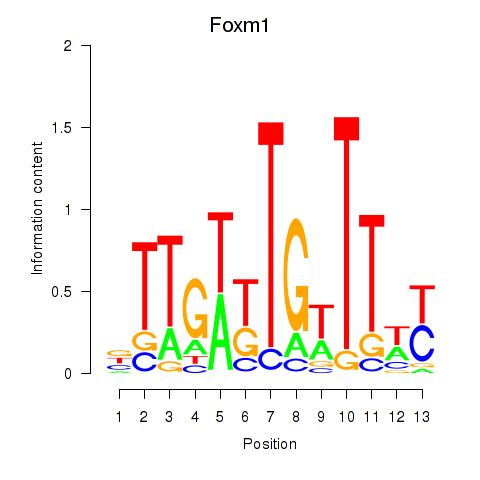
Results for Foxm1
Z-value: 1.10
Transcription factors associated with Foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxm1
|
ENSMUSG00000001517.15 | Foxm1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxm1 | mm39_v1_chr6_+_128339882_128340022 | -0.72 | 9.2e-07 | Click! |
Activity profile of Foxm1 motif
Sorted Z-values of Foxm1 motif
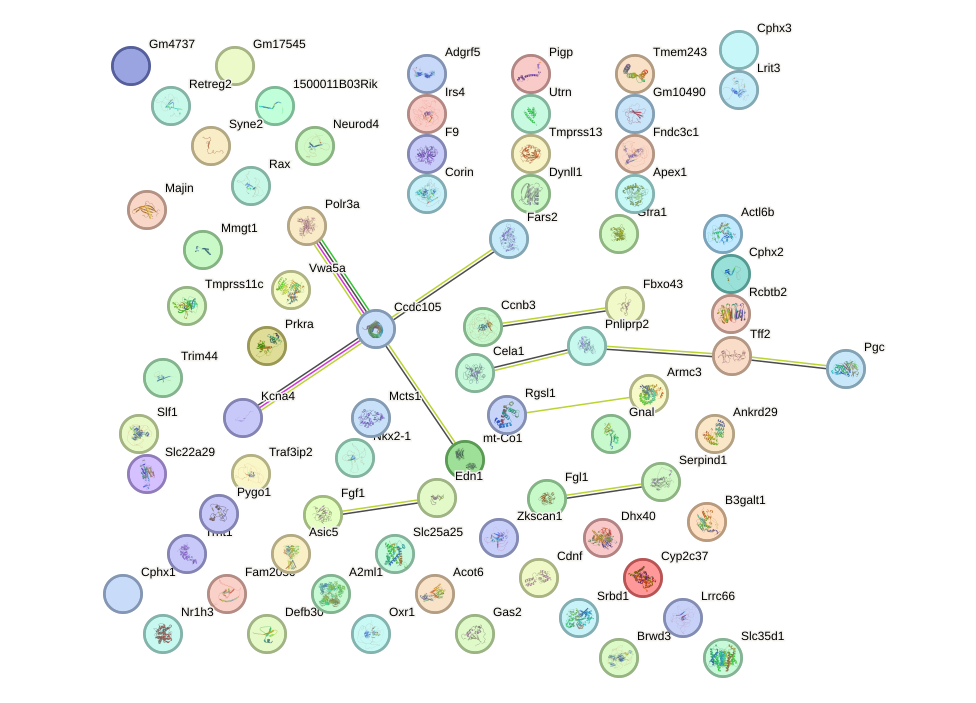
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxm1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_100579813 | 4.84 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr19_+_39980868 | 4.14 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr19_+_58748132 | 3.79 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr10_+_87694117 | 3.22 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chrX_+_59044796 | 3.01 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr17_-_31363245 | 2.95 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr16_-_45975440 | 2.88 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr19_-_8196196 | 2.65 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr2_+_67948057 | 2.25 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr5_+_9150691 | 2.16 |
ENSMUST00000115365.3
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr7_+_51537645 | 2.14 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr2_-_32314017 | 2.13 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr19_-_58443593 | 2.06 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr19_-_58443830 | 1.81 |
ENSMUST00000026076.14
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr9_+_22365586 | 1.68 |
ENSMUST00000168332.2
|
Gm17545
|
predicted gene, 17545 |
| chr3_+_20039775 | 1.45 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr19_-_58444336 | 1.35 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr2_-_91025492 | 1.26 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr3_+_81906768 | 1.21 |
ENSMUST00000107736.2
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr9_-_71070506 | 1.17 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr13_+_41040657 | 1.13 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr2_-_91025441 | 1.07 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr9_-_108140925 | 1.01 |
ENSMUST00000171412.7
ENSMUST00000195429.6 ENSMUST00000080435.9 |
Dag1
|
dystroglycan 1 |
| chr11_-_95966407 | 0.97 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr4_-_103072343 | 0.89 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr15_+_41615084 | 0.89 |
ENSMUST00000229511.2
ENSMUST00000229836.2 |
Oxr1
|
oxidation resistance 1 |
| chrM_+_5319 | 0.88 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr8_-_41668182 | 0.85 |
ENSMUST00000034003.5
|
Fgl1
|
fibrinogen-like protein 1 |
| chrX_+_141011695 | 0.85 |
ENSMUST00000112913.2
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chrX_-_55643429 | 0.84 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr13_+_24023428 | 0.79 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr13_+_36301331 | 0.76 |
ENSMUST00000021857.13
|
Fars2
|
phenylalanine-tRNA synthetase 2 (mitochondrial) |
| chr17_+_34524884 | 0.69 |
ENSMUST00000074557.11
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr2_+_107120934 | 0.68 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr17_+_34524841 | 0.66 |
ENSMUST00000235530.2
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr14_+_25942963 | 0.62 |
ENSMUST00000184016.3
|
Cphx1
|
cytoplasmic polyadenylated homeobox 1 |
| chr19_-_43879031 | 0.58 |
ENSMUST00000212048.2
|
Dnmbp
|
dynamin binding protein |
| chr18_-_39062201 | 0.58 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr14_+_73411249 | 0.55 |
ENSMUST00000166875.2
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr13_+_36301830 | 0.55 |
ENSMUST00000099582.4
ENSMUST00000224611.2 |
Fars2
|
phenylalanine-tRNA synthetase 2 (mitochondrial) |
| chr12_-_98540944 | 0.54 |
ENSMUST00000221305.2
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr16_+_17149235 | 0.53 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr13_+_24118417 | 0.52 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr2_-_132089543 | 0.51 |
ENSMUST00000110164.8
|
Tmem230
|
transmembrane protein 230 |
| chr1_+_75119419 | 0.51 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr10_-_12744025 | 0.50 |
ENSMUST00000219660.2
|
Utrn
|
utrophin |
| chr5_-_73789764 | 0.50 |
ENSMUST00000087177.4
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr19_-_32717166 | 0.49 |
ENSMUST00000235142.2
ENSMUST00000070210.6 ENSMUST00000236011.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr10_-_12743915 | 0.48 |
ENSMUST00000219584.2
|
Utrn
|
utrophin |
| chr17_-_86452564 | 0.48 |
ENSMUST00000095187.4
|
Srbd1
|
S1 RNA binding domain 1 |
| chr5_+_137551774 | 0.47 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr2_+_3514071 | 0.46 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr10_-_130116088 | 0.46 |
ENSMUST00000061571.5
|
Neurod4
|
neurogenic differentiation 4 |
| chr13_-_24118139 | 0.46 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr17_+_43700327 | 0.45 |
ENSMUST00000113599.2
ENSMUST00000224278.2 ENSMUST00000225466.2 |
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr9_+_72832904 | 0.44 |
ENSMUST00000038489.6
|
Pygo1
|
pygopus 1 |
| chr19_-_32717138 | 0.44 |
ENSMUST00000236985.2
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr10_+_39488930 | 0.44 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chrX_+_7775781 | 0.43 |
ENSMUST00000086274.3
|
Gm10490
|
predicted gene 10490 |
| chr5_+_137551790 | 0.42 |
ENSMUST00000136565.8
ENSMUST00000149292.8 ENSMUST00000125489.2 |
Actl6b
|
actin-like 6B |
| chr14_-_24537067 | 0.41 |
ENSMUST00000026322.9
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr2_-_102231208 | 0.41 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr11_-_86698484 | 0.40 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr4_+_143139571 | 0.39 |
ENSMUST00000155157.2
|
Pramel12
|
PRAME like 12 |
| chr19_+_6234392 | 0.38 |
ENSMUST00000025699.9
ENSMUST00000113528.2 |
Majin
|
membrane anchored junction protein |
| chr9_-_37623573 | 0.38 |
ENSMUST00000104875.2
|
Olfr160
|
olfactory receptor 160 |
| chr12_-_56583582 | 0.37 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr17_+_8591228 | 0.37 |
ENSMUST00000163578.8
|
T2
|
brachyury 2 |
| chr18_-_12429048 | 0.37 |
ENSMUST00000234212.2
|
Ankrd29
|
ankyrin repeat domain 29 |
| chr18_+_67266784 | 0.36 |
ENSMUST00000236918.2
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr4_-_42874178 | 0.36 |
ENSMUST00000107978.2
|
Fam205c
|
family with sequence similarity 205, member C |
| chr13_+_42454922 | 0.36 |
ENSMUST00000021796.9
|
Edn1
|
endothelin 1 |
| chr1_+_158189831 | 0.35 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr3_-_75389047 | 0.35 |
ENSMUST00000193989.4
ENSMUST00000203169.3 |
Wdr49
|
WD repeat domain 49 |
| chrX_-_140508177 | 0.35 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr4_-_42874197 | 0.34 |
ENSMUST00000055944.11
|
Fam205c
|
family with sequence similarity 205, member C |
| chr5_-_72661776 | 0.34 |
ENSMUST00000175766.8
ENSMUST00000177290.8 ENSMUST00000176974.2 ENSMUST00000167460.9 |
Corin
|
corin, serine peptidase |
| chr1_+_158190090 | 0.34 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chr8_-_37200051 | 0.34 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr5_+_31684331 | 0.33 |
ENSMUST00000114533.9
ENSMUST00000202214.4 ENSMUST00000201858.4 ENSMUST00000202950.4 |
Slc4a1ap
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr4_+_143139490 | 0.32 |
ENSMUST00000132915.2
ENSMUST00000037356.8 |
Pramel12
|
PRAME like 12 |
| chr3_-_59102517 | 0.32 |
ENSMUST00000200095.2
|
Gpr87
|
G protein-coupled receptor 87 |
| chr16_-_94172691 | 0.31 |
ENSMUST00000232294.2
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr6_-_128558560 | 0.31 |
ENSMUST00000060574.9
|
A2ml1
|
alpha-2-macroglobulin like 1 |
| chr10_-_20600797 | 0.31 |
ENSMUST00000020165.14
|
Pde7b
|
phosphodiesterase 7B |
| chr17_+_48037758 | 0.31 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr14_-_63287340 | 0.30 |
ENSMUST00000111209.2
ENSMUST00000111208.2 |
Defb30
|
defensin beta 30 |
| chrX_+_37689503 | 0.30 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr1_+_158189900 | 0.30 |
ENSMUST00000170718.7
|
Astn1
|
astrotactin 1 |
| chr9_+_38629560 | 0.29 |
ENSMUST00000001544.12
ENSMUST00000118144.8 |
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr1_-_153719876 | 0.29 |
ENSMUST00000238926.2
ENSMUST00000124558.10 |
Rgsl1
|
regulator of G-protein signaling like 1 |
| chr12_+_84147571 | 0.28 |
ENSMUST00000222921.2
|
Acot6
|
acyl-CoA thioesterase 6 |
| chr18_-_66072119 | 0.28 |
ENSMUST00000025396.5
|
Rax
|
retina and anterior neural fold homeobox |
| chrX_-_105528821 | 0.27 |
ENSMUST00000039447.14
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chrX_-_107877909 | 0.27 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr8_-_58249608 | 0.27 |
ENSMUST00000204067.3
|
Galntl6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr12_+_76118790 | 0.27 |
ENSMUST00000131480.8
|
Syne2
|
spectrin repeat containing, nuclear envelope 2 |
| chr15_-_36164963 | 0.26 |
ENSMUST00000227793.2
|
Fbxo43
|
F-box protein 43 |
| chr13_-_77279395 | 0.26 |
ENSMUST00000159300.8
|
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr12_-_85143852 | 0.26 |
ENSMUST00000177289.9
|
Prox2
|
prospero homeobox 2 |
| chrX_-_6907858 | 0.26 |
ENSMUST00000115752.8
|
Ccnb3
|
cyclin B3 |
| chr3_-_129597679 | 0.25 |
ENSMUST00000185462.7
ENSMUST00000179187.2 |
Lrit3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr5_+_138083345 | 0.25 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr11_-_100244866 | 0.25 |
ENSMUST00000173630.8
|
Hap1
|
huntingtin-associated protein 1 |
| chr10_-_78588884 | 0.24 |
ENSMUST00000105383.4
|
Ccdc105
|
coiled-coil domain containing 105 |
| chr6_+_106746138 | 0.23 |
ENSMUST00000205163.2
|
Trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr2_+_19204076 | 0.23 |
ENSMUST00000114640.9
ENSMUST00000049255.7 |
Armc3
|
armadillo repeat containing 3 |
| chr11_-_69706124 | 0.23 |
ENSMUST00000210714.2
|
Gm39566
|
predicted gene, 39566 |
| chr9_+_45230370 | 0.23 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr7_-_104570689 | 0.22 |
ENSMUST00000217177.2
|
Olfr667
|
olfactory receptor 667 |
| chr6_+_32027216 | 0.22 |
ENSMUST00000031778.6
|
1700012A03Rik
|
RIKEN cDNA 1700012A03 gene |
| chr19_+_25649767 | 0.22 |
ENSMUST00000053068.7
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr13_-_19521337 | 0.22 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chr2_+_153621851 | 0.22 |
ENSMUST00000126656.4
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr5_-_125467063 | 0.22 |
ENSMUST00000136312.2
|
Ubc
|
ubiquitin C |
| chr3_+_33853941 | 0.21 |
ENSMUST00000099153.10
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr10_+_90665639 | 0.20 |
ENSMUST00000179337.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_-_42442130 | 0.20 |
ENSMUST00000216650.2
|
Olfr458
|
olfactory receptor 458 |
| chr14_+_76192449 | 0.20 |
ENSMUST00000050120.4
|
Kctd4
|
potassium channel tetramerisation domain containing 4 |
| chr8_-_25086976 | 0.20 |
ENSMUST00000033956.7
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr11_-_99313078 | 0.20 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr4_-_143026033 | 0.19 |
ENSMUST00000119654.2
|
Pdpn
|
podoplanin |
| chr10_-_5019044 | 0.19 |
ENSMUST00000095899.5
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr4_+_98434930 | 0.18 |
ENSMUST00000102792.10
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr16_-_58437336 | 0.18 |
ENSMUST00000099663.3
|
Ftdc1
|
ferritin domain containing 1 |
| chr10_-_81066607 | 0.18 |
ENSMUST00000047408.6
|
Atcay
|
ataxia, cerebellar, Cayman type |
| chr18_-_66155651 | 0.18 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr1_-_79417732 | 0.18 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chrY_-_9132561 | 0.17 |
ENSMUST00000171947.3
|
Gm21292
|
predicted gene, 21292 |
| chr19_+_5416769 | 0.17 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr11_+_93935156 | 0.17 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr2_+_10377237 | 0.17 |
ENSMUST00000114864.9
ENSMUST00000116594.9 ENSMUST00000041105.7 |
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr4_+_57742077 | 0.17 |
ENSMUST00000107600.8
|
Pakap
|
paralemmin A kinase anchor protein |
| chr2_+_135894099 | 0.17 |
ENSMUST00000144403.8
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr12_-_85317359 | 0.16 |
ENSMUST00000166821.8
ENSMUST00000019378.8 ENSMUST00000220854.2 |
Mlh3
|
mutL homolog 3 |
| chr9_+_20729784 | 0.16 |
ENSMUST00000066387.6
|
Rdh8
|
retinol dehydrogenase 8 |
| chr7_+_99512701 | 0.16 |
ENSMUST00000207855.2
|
Xrra1
|
X-ray radiation resistance associated 1 |
| chr13_+_83720484 | 0.16 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr5_-_86666408 | 0.15 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chrY_-_79161056 | 0.15 |
ENSMUST00000179922.2
|
Gm20917
|
predicted gene, 20917 |
| chr7_-_103390529 | 0.15 |
ENSMUST00000051346.5
|
Olfr629
|
olfactory receptor 629 |
| chr6_+_145879839 | 0.15 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr1_-_138770167 | 0.15 |
ENSMUST00000112026.4
ENSMUST00000019374.14 |
Lhx9
|
LIM homeobox protein 9 |
| chr2_+_136374186 | 0.15 |
ENSMUST00000121717.2
|
Ankef1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr17_+_28749780 | 0.14 |
ENSMUST00000233923.2
|
Armc12
|
armadillo repeat containing 12 |
| chr19_+_56450062 | 0.14 |
ENSMUST00000178590.9
ENSMUST00000039666.8 |
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr4_-_15149755 | 0.14 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr10_+_23727325 | 0.13 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr19_+_56450085 | 0.13 |
ENSMUST00000225909.2
|
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr2_+_14179324 | 0.12 |
ENSMUST00000077517.9
|
Tmem236
|
transmembrane protein 236 |
| chr2_+_96148418 | 0.12 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chrY_+_21242966 | 0.12 |
ENSMUST00000179095.2
|
Gm20865
|
predicted gene, 20865 |
| chrX_+_111404963 | 0.11 |
ENSMUST00000026602.9
ENSMUST00000113412.3 |
2010106E10Rik
|
RIKEN cDNA 2010106E10 gene |
| chr3_-_152373997 | 0.10 |
ENSMUST00000045262.11
|
Ak5
|
adenylate kinase 5 |
| chr7_+_75351713 | 0.10 |
ENSMUST00000207239.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr2_+_103926140 | 0.10 |
ENSMUST00000168176.8
ENSMUST00000040423.12 |
Cd59a
|
CD59a antigen |
| chr11_+_93935066 | 0.10 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr7_+_5037117 | 0.10 |
ENSMUST00000076791.4
|
4632433K11Rik
|
RIKEN cDNA 4632433K11 gene |
| chr2_-_73283010 | 0.09 |
ENSMUST00000151939.2
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_-_127782735 | 0.09 |
ENSMUST00000208183.3
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr3_+_133942244 | 0.09 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr6_+_70680515 | 0.09 |
ENSMUST00000103404.2
|
Igkv3-1
|
immunoglobulin kappa variable 3-1 |
| chr5_+_135661472 | 0.09 |
ENSMUST00000043707.7
|
Rhbdd2
|
rhomboid domain containing 2 |
| chr4_-_144134967 | 0.09 |
ENSMUST00000123854.2
ENSMUST00000030326.10 |
Pramel13
|
PRAME like 13 |
| chr11_+_120489247 | 0.08 |
ENSMUST00000026128.10
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr19_+_13477910 | 0.07 |
ENSMUST00000217001.2
|
Olfr1477
|
olfactory receptor 1477 |
| chr7_+_27510238 | 0.07 |
ENSMUST00000080175.7
|
Zfp626
|
zinc finger protein 626 |
| chr10_-_129951252 | 0.07 |
ENSMUST00000213145.2
|
Olfr823
|
olfactory receptor 823 |
| chr10_+_127685785 | 0.06 |
ENSMUST00000077530.3
|
Rdh19
|
retinol dehydrogenase 19 |
| chr3_-_80710097 | 0.06 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr5_-_125467240 | 0.06 |
ENSMUST00000156249.2
|
Ubc
|
ubiquitin C |
| chr13_+_83720457 | 0.06 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chrX_+_159551009 | 0.06 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr7_+_87927293 | 0.06 |
ENSMUST00000032779.12
ENSMUST00000131108.9 ENSMUST00000128791.2 |
Ctsc
|
cathepsin C |
| chr17_+_26042600 | 0.05 |
ENSMUST00000026833.6
|
Wdr24
|
WD repeat domain 24 |
| chr19_+_13560875 | 0.05 |
ENSMUST00000208104.3
ENSMUST00000216014.3 ENSMUST00000216369.2 ENSMUST00000217451.3 ENSMUST00000215567.3 |
Olfr1484
|
olfactory receptor 1484 |
| chr2_+_57887896 | 0.05 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr3_+_99048379 | 0.05 |
ENSMUST00000004343.7
|
Wars2
|
tryptophanyl tRNA synthetase 2 (mitochondrial) |
| chr14_+_53100756 | 0.05 |
ENSMUST00000103616.5
ENSMUST00000186370.2 |
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr12_-_87691390 | 0.05 |
ENSMUST00000220528.2
|
Ubl5b
|
ubiquitin-like 5B |
| chr1_+_179788037 | 0.04 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_-_99512558 | 0.04 |
ENSMUST00000207137.2
ENSMUST00000207063.2 ENSMUST00000207580.2 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr9_+_37714354 | 0.04 |
ENSMUST00000215287.2
|
Olfr876
|
olfactory receptor 876 |
| chr4_+_98435155 | 0.04 |
ENSMUST00000030290.8
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr6_-_13871475 | 0.04 |
ENSMUST00000139231.2
|
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr11_+_93934940 | 0.04 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr2_+_65676111 | 0.02 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr9_+_19624125 | 0.02 |
ENSMUST00000077023.4
|
Olfr857
|
olfactory receptor 857 |
| chr2_+_135501605 | 0.02 |
ENSMUST00000134310.8
|
Plcb4
|
phospholipase C, beta 4 |
| chr11_-_58521327 | 0.01 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr10_-_13068900 | 0.01 |
ENSMUST00000019950.6
|
Ltv1
|
LTV1 ribosome biogenesis factor |
| chrX_+_94942639 | 0.01 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr6_+_43149074 | 0.01 |
ENSMUST00000216179.2
|
Olfr13
|
olfactory receptor 13 |
| chr5_+_67125759 | 0.01 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr11_+_74150634 | 0.00 |
ENSMUST00000217016.2
ENSMUST00000214303.2 |
Olfr406
|
olfactory receptor 406 |
| chr8_-_21817031 | 0.00 |
ENSMUST00000098890.4
|
Defa29
|
defensin, alpha, 29 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.8 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.6 | 2.3 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.5 | 3.2 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.4 | 2.9 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.4 | 6.0 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 0.3 | 1.0 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.3 | 1.2 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.3 | 1.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 3.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 0.3 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.0 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 4.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.4 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.2 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.3 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.1 | 0.3 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 0.4 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 2.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0032899 | regulation of neurotrophin production(GO:0032899) positive regulation of neurotrophin production(GO:0032901) |
| 0.0 | 0.4 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.3 | GO:1990166 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 3.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 5.2 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 2.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 3.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.0 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 1.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 5.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 2.3 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 1.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 3.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.2 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.2 | 1.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 2.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 3.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 2.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 4.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.9 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.8 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 3.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 8.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.7 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 5.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 7.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 5.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 3.8 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |