Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
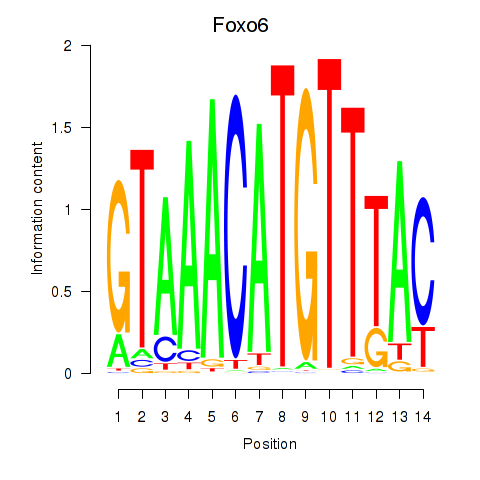
Results for Foxo6
Z-value: 1.36
Transcription factors associated with Foxo6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo6
|
ENSMUSG00000052135.9 | Foxo6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo6 | mm39_v1_chr4_-_120144546_120144546 | 0.66 | 1.4e-05 | Click! |
Activity profile of Foxo6 motif
Sorted Z-values of Foxo6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxo6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87684299 | 13.87 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr16_-_36228798 | 12.80 |
ENSMUST00000023619.8
|
Stfa2
|
stefin A2 |
| chr15_-_74983786 | 11.11 |
ENSMUST00000191451.2
ENSMUST00000100542.10 |
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chr11_-_11977958 | 9.29 |
ENSMUST00000109654.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr9_+_98372575 | 8.05 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr15_+_103362195 | 6.37 |
ENSMUST00000047405.9
|
Nckap1l
|
NCK associated protein 1 like |
| chr2_+_118428690 | 4.54 |
ENSMUST00000038341.8
|
Bub1b
|
BUB1B, mitotic checkpoint serine/threonine kinase |
| chr6_-_41012435 | 3.96 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr4_+_134238310 | 3.89 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr5_+_110478558 | 3.34 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr14_-_70945434 | 3.33 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr2_-_25498459 | 3.14 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr12_-_91351177 | 3.04 |
ENSMUST00000141429.8
|
Cep128
|
centrosomal protein 128 |
| chr5_-_117527094 | 3.04 |
ENSMUST00000111953.2
ENSMUST00000086461.13 |
Rfc5
|
replication factor C (activator 1) 5 |
| chr19_+_8870362 | 2.98 |
ENSMUST00000096249.7
|
Ints5
|
integrator complex subunit 5 |
| chr18_-_32044877 | 2.95 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr7_+_28140352 | 2.92 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr2_+_129435115 | 2.91 |
ENSMUST00000099113.10
ENSMUST00000103202.10 |
Sirpa
|
signal-regulatory protein alpha |
| chr16_+_16964801 | 2.68 |
ENSMUST00000232479.2
ENSMUST00000232344.2 ENSMUST00000069064.7 |
Ydjc
|
YdjC homolog (bacterial) |
| chr3_+_84573499 | 2.63 |
ENSMUST00000107682.2
|
Tmem154
|
transmembrane protein 154 |
| chr5_-_65548723 | 2.60 |
ENSMUST00000120094.5
ENSMUST00000127874.6 ENSMUST00000057885.13 |
Rpl9
|
ribosomal protein L9 |
| chr2_+_91480513 | 2.42 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr14_+_31888061 | 2.40 |
ENSMUST00000164341.2
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chrX_+_133486391 | 2.31 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr2_+_129434738 | 2.13 |
ENSMUST00000153491.8
ENSMUST00000161620.8 ENSMUST00000179001.8 |
Sirpa
|
signal-regulatory protein alpha |
| chr7_+_28140450 | 2.09 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr11_+_68986043 | 2.06 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr17_+_48653493 | 1.82 |
ENSMUST00000113237.4
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr6_-_119925387 | 1.75 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chrX_-_73325318 | 1.68 |
ENSMUST00000239458.2
ENSMUST00000019232.10 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr12_-_113802603 | 1.65 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr2_+_163916042 | 1.60 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr13_-_96803216 | 1.58 |
ENSMUST00000170287.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr2_-_92876398 | 1.55 |
ENSMUST00000111272.3
ENSMUST00000178666.8 ENSMUST00000147339.3 |
Prdm11
|
PR domain containing 11 |
| chrX_-_73325204 | 1.55 |
ENSMUST00000114189.10
ENSMUST00000119361.4 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr15_+_75140241 | 1.53 |
ENSMUST00000023247.13
|
Ly6f
|
lymphocyte antigen 6 complex, locus F |
| chr15_+_52575796 | 1.52 |
ENSMUST00000037115.9
|
Med30
|
mediator complex subunit 30 |
| chr17_-_33879224 | 1.51 |
ENSMUST00000130946.8
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr6_+_57679621 | 1.46 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr14_-_49482846 | 1.43 |
ENSMUST00000227113.2
ENSMUST00000130853.2 ENSMUST00000228936.2 ENSMUST00000022398.15 |
ccdc198
|
coiled-coil domain containing 198 |
| chr15_-_74855264 | 1.40 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr2_-_84255602 | 1.38 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr2_+_129434834 | 1.33 |
ENSMUST00000103203.8
|
Sirpa
|
signal-regulatory protein alpha |
| chr12_-_102390000 | 1.31 |
ENSMUST00000110020.8
|
Lgmn
|
legumain |
| chr1_+_171097891 | 1.21 |
ENSMUST00000064272.10
ENSMUST00000151863.8 ENSMUST00000141999.8 ENSMUST00000111313.10 ENSMUST00000126699.4 |
B4galt3
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 3 |
| chr6_+_122929410 | 1.15 |
ENSMUST00000117173.8
|
Clec4a3
|
C-type lectin domain family 4, member a3 |
| chr13_+_76727787 | 1.06 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr4_-_136626073 | 1.06 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr1_-_38704028 | 0.99 |
ENSMUST00000039827.14
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr18_+_37884653 | 0.98 |
ENSMUST00000194928.2
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr7_-_122969064 | 0.97 |
ENSMUST00000207010.2
ENSMUST00000167309.8 ENSMUST00000205262.2 ENSMUST00000106442.9 ENSMUST00000098060.5 |
Arhgap17
|
Rho GTPase activating protein 17 |
| chr6_+_57679455 | 0.97 |
ENSMUST00000072954.8
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr2_-_130928658 | 0.96 |
ENSMUST00000028794.10
ENSMUST00000110227.8 ENSMUST00000110226.2 |
Siglec1
|
sialic acid binding Ig-like lectin 1, sialoadhesin |
| chr17_+_25690538 | 0.94 |
ENSMUST00000234449.2
ENSMUST00000025002.3 ENSMUST00000235033.2 |
Tekt4
|
tektin 4 |
| chr9_+_109865810 | 0.92 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr9_-_21982637 | 0.92 |
ENSMUST00000179605.9
ENSMUST00000043922.7 |
Zfp653
|
zinc finger protein 653 |
| chr19_-_17316906 | 0.91 |
ENSMUST00000169897.2
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr7_+_28508220 | 0.89 |
ENSMUST00000172529.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr10_+_39296005 | 0.86 |
ENSMUST00000157009.8
|
Fyn
|
Fyn proto-oncogene |
| chr2_-_85966272 | 0.84 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr16_+_51870001 | 0.82 |
ENSMUST00000227756.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr12_+_108376801 | 0.81 |
ENSMUST00000054955.14
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chrX_+_105964224 | 0.78 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr6_-_129449739 | 0.77 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr3_+_103875574 | 0.75 |
ENSMUST00000063717.14
ENSMUST00000055425.15 ENSMUST00000123611.8 ENSMUST00000090685.11 |
Phtf1
|
putative homeodomain transcription factor 1 |
| chr6_+_87405968 | 0.69 |
ENSMUST00000032125.7
|
Bmp10
|
bone morphogenetic protein 10 |
| chr17_+_64203017 | 0.67 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr4_-_62389098 | 0.67 |
ENSMUST00000135811.2
ENSMUST00000120095.8 ENSMUST00000030087.14 ENSMUST00000107452.8 ENSMUST00000155522.8 |
Wdr31
|
WD repeat domain 31 |
| chr4_-_138858340 | 0.63 |
ENSMUST00000143971.2
|
Micos10
|
mitochondrial contact site and cristae organizing system subunit 10 |
| chr3_-_88455556 | 0.60 |
ENSMUST00000131775.2
ENSMUST00000008745.13 |
Rab25
|
RAB25, member RAS oncogene family |
| chr12_+_108376884 | 0.58 |
ENSMUST00000109857.8
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr13_+_76727799 | 0.54 |
ENSMUST00000109589.3
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr4_+_108336164 | 0.51 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr2_-_155199300 | 0.50 |
ENSMUST00000165234.2
ENSMUST00000077626.13 |
Pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr11_-_59727752 | 0.48 |
ENSMUST00000156837.2
|
Cops3
|
COP9 signalosome subunit 3 |
| chr7_+_84502761 | 0.44 |
ENSMUST00000217039.3
ENSMUST00000211582.2 |
Olfr291
|
olfactory receptor 291 |
| chr9_+_38340751 | 0.43 |
ENSMUST00000216502.3
ENSMUST00000216644.2 |
Olfr901
|
olfactory receptor 901 |
| chr12_-_115832846 | 0.43 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr2_-_86061745 | 0.39 |
ENSMUST00000216056.2
|
Olfr1047
|
olfactory receptor 1047 |
| chr6_-_57821483 | 0.38 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr15_+_41573995 | 0.37 |
ENSMUST00000229769.2
|
Oxr1
|
oxidation resistance 1 |
| chr6_+_42885812 | 0.35 |
ENSMUST00000216408.2
|
Olfr447
|
olfactory receptor 447 |
| chr4_+_80752360 | 0.33 |
ENSMUST00000133655.8
ENSMUST00000006151.13 |
Tyrp1
|
tyrosinase-related protein 1 |
| chr17_-_23783063 | 0.33 |
ENSMUST00000095606.5
|
Zfp213
|
zinc finger protein 213 |
| chr15_+_100052260 | 0.32 |
ENSMUST00000023768.14
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr17_-_80885197 | 0.31 |
ENSMUST00000234602.2
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr8_+_26401698 | 0.31 |
ENSMUST00000120653.8
ENSMUST00000126226.2 |
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr1_-_65158717 | 0.31 |
ENSMUST00000144760.3
|
Gm28845
|
predicted gene 28845 |
| chr8_+_71176713 | 0.30 |
ENSMUST00000034307.14
ENSMUST00000239435.2 ENSMUST00000239487.2 ENSMUST00000110095.3 |
Pde4c
|
phosphodiesterase 4C, cAMP specific |
| chr11_+_46126274 | 0.30 |
ENSMUST00000060185.3
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr14_+_51648277 | 0.29 |
ENSMUST00000159674.4
ENSMUST00000163019.9 ENSMUST00000233394.2 ENSMUST00000232951.2 ENSMUST00000232797.2 ENSMUST00000233766.2 ENSMUST00000233848.2 ENSMUST00000233280.2 ENSMUST00000233920.2 ENSMUST00000233126.2 ENSMUST00000233651.2 |
Vmn2r88
|
vomeronasal 2, receptor 88 |
| chr10_+_23672842 | 0.24 |
ENSMUST00000119597.8
ENSMUST00000179321.8 ENSMUST00000133289.2 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr18_-_60781365 | 0.24 |
ENSMUST00000143275.3
|
Synpo
|
synaptopodin |
| chr7_-_102566717 | 0.19 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr2_+_36342599 | 0.17 |
ENSMUST00000072854.2
|
Olfr340
|
olfactory receptor 340 |
| chr13_-_22375311 | 0.16 |
ENSMUST00000238109.2
|
Vmn1r192
|
vomeronasal 1 receptor 192 |
| chr16_-_88360037 | 0.14 |
ENSMUST00000049697.5
|
Cldn8
|
claudin 8 |
| chr11_+_87744033 | 0.13 |
ENSMUST00000038196.7
|
Mks1
|
MKS transition zone complex subunit 1 |
| chr18_-_12438695 | 0.13 |
ENSMUST00000122408.8
ENSMUST00000234183.2 ENSMUST00000142066.2 ENSMUST00000118525.9 |
Ankrd29
|
ankyrin repeat domain 29 |
| chr11_-_121563894 | 0.12 |
ENSMUST00000067399.14
ENSMUST00000062654.8 |
B3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr5_+_88150390 | 0.12 |
ENSMUST00000189633.7
ENSMUST00000031211.6 |
Smr3a
|
submaxillary gland androgen regulated protein 3A |
| chrX_-_123045649 | 0.11 |
ENSMUST00000094491.5
|
Vmn2r121
|
vomeronasal 2, receptor 121 |
| chr10_+_33913593 | 0.11 |
ENSMUST00000095758.3
|
Trappc3l
|
trafficking protein particle complex 3 like |
| chr14_+_53478202 | 0.10 |
ENSMUST00000179583.3
|
Trav12n-3
|
T cell receptor alpha variable 12N-3 |
| chr12_-_115019136 | 0.10 |
ENSMUST00000103519.2
ENSMUST00000192724.2 |
Ighv1-49
|
immunoglobulin heavy variable 1-49 |
| chr11_+_29413734 | 0.10 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr3_+_103875261 | 0.09 |
ENSMUST00000117150.8
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr4_+_43851565 | 0.08 |
ENSMUST00000107860.3
|
Olfr155
|
olfactory receptor 155 |
| chr14_+_65842716 | 0.08 |
ENSMUST00000079469.7
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr4_+_62204678 | 0.07 |
ENSMUST00000084530.9
|
Slc31a2
|
solute carrier family 31, member 2 |
| chr1_+_179788675 | 0.07 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr17_-_31496301 | 0.06 |
ENSMUST00000235144.2
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr4_-_156134922 | 0.05 |
ENSMUST00000184684.8
|
Ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr5_+_115417725 | 0.04 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr2_-_111820618 | 0.02 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr12_-_98225676 | 0.01 |
ENSMUST00000021390.9
|
Galc
|
galactosylceramidase |
| chr6_-_54949587 | 0.01 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr10_+_129072073 | 0.00 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chr6_-_68784692 | 0.00 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr6_-_130044234 | 0.00 |
ENSMUST00000119096.2
|
Klra4
|
killer cell lectin-like receptor, subfamily A, member 4 |
| chr10_-_130333265 | 0.00 |
ENSMUST00000164227.4
|
Vmn2r87
|
vomeronasal 2, receptor 87 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.9 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.7 | 5.0 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.6 | 6.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 1.1 | 3.3 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.9 | 9.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.9 | 4.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.9 | 8.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.6 | 2.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.6 | 1.8 | GO:0032499 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.4 | 1.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.4 | 2.4 | GO:0032439 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.4 | 1.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 1.7 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.3 | 1.4 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.3 | 2.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.3 | 3.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.9 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 1.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 3.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 2.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.3 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.9 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.7 | GO:0038095 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.8 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.9 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) positive regulation of sarcomere organization(GO:0060298) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.1 | 6.4 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.1 | 12.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.9 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 1.0 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 3.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.6 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 2.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 5.3 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.4 | GO:1902083 | cellular response to hydroperoxide(GO:0071447) negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.8 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 1.0 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 1.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:1990403 | embryonic brain development(GO:1990403) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.7 | 3.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 3.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 6.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 2.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 4.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 3.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 2.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 4.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 12.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 3.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 2.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.5 | 1.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.4 | 8.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 5.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 3.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 3.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 3.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 1.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.2 | 1.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 13.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 0.9 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 16.5 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 1.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 12.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.5 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 6.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 1.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 2.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 9.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 8.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.4 | 6.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 6.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 9.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 4.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 1.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 4.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 3.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 3.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |