Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxp1_Foxj2
Z-value: 3.27
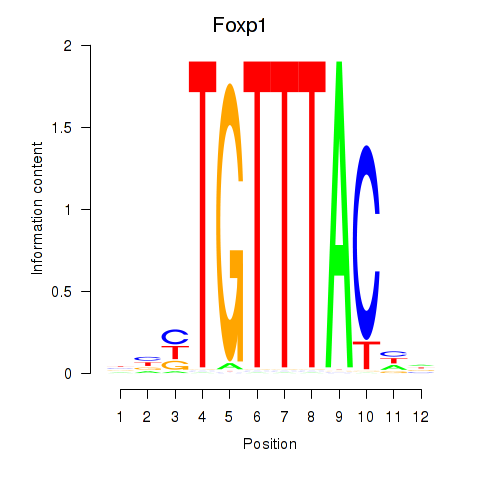
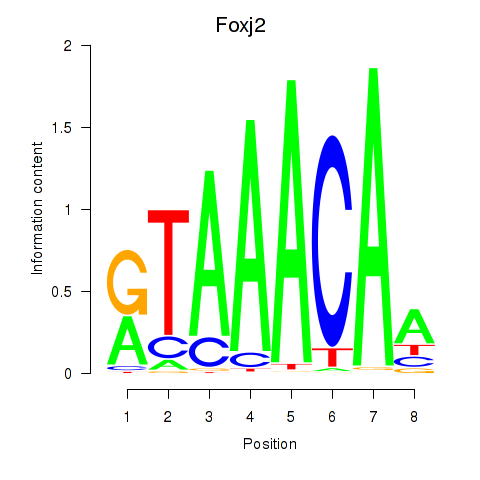
Transcription factors associated with Foxp1_Foxj2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxp1
|
ENSMUSG00000030067.18 | Foxp1 |
|
Foxj2
|
ENSMUSG00000003154.16 | Foxj2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxp1 | mm39_v1_chr6_-_99005835_99005903 | -0.66 | 1.0e-05 | Click! |
| Foxj2 | mm39_v1_chr6_+_122796847_122796897 | -0.60 | 1.1e-04 | Click! |
Activity profile of Foxp1_Foxj2 motif
Sorted Z-values of Foxp1_Foxj2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxp1_Foxj2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_130589349 | 20.50 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr1_-_130589321 | 20.35 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr6_+_121983720 | 19.14 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr5_-_145816774 | 18.99 |
ENSMUST00000035918.8
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr3_+_138121245 | 14.72 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr14_+_55797934 | 12.85 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr17_-_12894716 | 12.61 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr14_-_52150804 | 12.23 |
ENSMUST00000004673.15
ENSMUST00000111632.5 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr14_-_52151026 | 12.07 |
ENSMUST00000228164.2
|
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr9_-_48516447 | 11.78 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr8_-_25066313 | 11.66 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr16_+_22739028 | 11.10 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr19_+_30210320 | 10.94 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chrX_+_138701544 | 10.71 |
ENSMUST00000054889.4
|
Cldn2
|
claudin 2 |
| chr8_-_93806593 | 10.53 |
ENSMUST00000109582.3
|
Ces1b
|
carboxylesterase 1B |
| chr14_-_52151537 | 10.50 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr2_+_67948057 | 10.50 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr8_-_122671588 | 10.38 |
ENSMUST00000057653.8
|
Car5a
|
carbonic anhydrase 5a, mitochondrial |
| chr10_+_93324624 | 10.03 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase |
| chr15_-_78352801 | 9.92 |
ENSMUST00000229124.2
ENSMUST00000230226.2 ENSMUST00000017086.5 |
Tmprss6
|
transmembrane serine protease 6 |
| chr14_+_55798362 | 9.72 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr15_+_6474808 | 9.66 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr14_+_55798517 | 9.35 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr12_-_103623418 | 9.11 |
ENSMUST00000044159.7
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr2_+_58644922 | 9.10 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr6_+_121323577 | 8.81 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr11_-_86884507 | 8.72 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr19_-_39451509 | 8.70 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr10_+_127734384 | 8.69 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr10_+_128089965 | 8.67 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr1_-_180023467 | 8.64 |
ENSMUST00000161746.2
ENSMUST00000160879.7 |
Coq8a
|
coenzyme Q8A |
| chr15_+_99290832 | 8.62 |
ENSMUST00000160635.8
ENSMUST00000161250.8 ENSMUST00000229392.2 ENSMUST00000161778.8 |
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr16_+_22739191 | 8.56 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr1_-_180023518 | 8.51 |
ENSMUST00000162769.8
ENSMUST00000161379.2 ENSMUST00000027766.13 ENSMUST00000161814.8 |
Coq8a
|
coenzyme Q8A |
| chr16_+_22738987 | 8.49 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr15_+_99290763 | 8.44 |
ENSMUST00000023749.15
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_+_133280680 | 8.25 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr10_-_115197775 | 8.04 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr17_-_46749370 | 7.96 |
ENSMUST00000087012.7
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr17_-_34962823 | 7.92 |
ENSMUST00000069507.9
|
C4b
|
complement component 4B (Chido blood group) |
| chr12_-_84497718 | 7.90 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr15_+_99291100 | 7.86 |
ENSMUST00000159209.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr7_+_140343652 | 7.77 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr15_+_3300249 | 7.65 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr2_+_43445333 | 7.58 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr8_-_94006345 | 7.23 |
ENSMUST00000034178.9
|
Ces1f
|
carboxylesterase 1F |
| chr11_+_69945157 | 6.89 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr19_-_40175709 | 6.85 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr19_+_7034149 | 6.84 |
ENSMUST00000040261.7
|
Macrod1
|
mono-ADP ribosylhydrolase 1 |
| chr10_+_87695352 | 6.80 |
ENSMUST00000121952.8
ENSMUST00000126490.8 |
Igf1
|
insulin-like growth factor 1 |
| chr18_+_51250748 | 6.70 |
ENSMUST00000116639.4
|
Prr16
|
proline rich 16 |
| chr17_-_46749320 | 6.64 |
ENSMUST00000233575.2
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr1_-_184543367 | 6.58 |
ENSMUST00000048462.13
ENSMUST00000110992.9 |
Mtarc1
|
mitochondrial amidoxime reducing component 1 |
| chr2_-_110144869 | 6.50 |
ENSMUST00000133608.2
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chrX_+_7588505 | 6.35 |
ENSMUST00000207675.2
ENSMUST00000116633.9 ENSMUST00000208996.2 ENSMUST00000144148.4 ENSMUST00000125991.9 ENSMUST00000148624.8 |
Wdr45
|
WD repeat domain 45 |
| chr2_+_43445359 | 6.29 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr3_+_146276147 | 6.21 |
ENSMUST00000199489.5
|
Uox
|
urate oxidase |
| chr10_-_18110682 | 6.19 |
ENSMUST00000052648.9
ENSMUST00000080860.13 ENSMUST00000173243.8 |
Ccdc28a
|
coiled-coil domain containing 28A |
| chr7_+_46401214 | 6.15 |
ENSMUST00000210769.2
ENSMUST00000210272.2 ENSMUST00000075982.4 |
Saa2
|
serum amyloid A 2 |
| chr18_-_39051695 | 6.11 |
ENSMUST00000040647.11
|
Fgf1
|
fibroblast growth factor 1 |
| chr10_+_111342147 | 6.08 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1 |
| chr1_-_192923816 | 6.04 |
ENSMUST00000160929.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr18_-_3281089 | 5.98 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr1_-_179373826 | 5.88 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr10_+_87695886 | 5.87 |
ENSMUST00000062862.13
|
Igf1
|
insulin-like growth factor 1 |
| chrX_+_7588453 | 5.83 |
ENSMUST00000043045.10
ENSMUST00000207386.2 ENSMUST00000116634.9 ENSMUST00000208072.2 ENSMUST00000207589.2 ENSMUST00000208618.2 ENSMUST00000208443.2 ENSMUST00000207541.2 ENSMUST00000208528.2 ENSMUST00000115689.10 ENSMUST00000131077.9 ENSMUST00000115688.8 ENSMUST00000208156.2 |
Wdr45
Gm45208
|
WD repeat domain 45 predicted gene 45208 |
| chr1_+_88030951 | 5.81 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr18_-_61669641 | 5.76 |
ENSMUST00000237557.2
ENSMUST00000171629.3 |
Arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr3_+_146302832 | 5.76 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr2_+_173579285 | 5.72 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr2_+_58645189 | 5.61 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr5_+_31079177 | 5.58 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr12_-_72675624 | 5.53 |
ENSMUST00000208039.2
ENSMUST00000207585.2 |
Gm4756
|
predicted gene 4756 |
| chr8_-_85500010 | 5.52 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr11_+_114741948 | 5.52 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr4_+_144619397 | 5.49 |
ENSMUST00000105744.8
ENSMUST00000171001.8 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr3_+_85946145 | 5.49 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr9_-_121745354 | 5.45 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr10_-_89369432 | 5.39 |
ENSMUST00000105297.2
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_+_146016064 | 5.39 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr7_-_119122681 | 5.30 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr6_+_134807097 | 5.29 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr8_+_105996419 | 5.29 |
ENSMUST00000036127.9
ENSMUST00000163734.9 |
Hsf4
|
heat shock transcription factor 4 |
| chr16_+_19916292 | 5.26 |
ENSMUST00000023509.5
ENSMUST00000232088.2 ENSMUST00000231842.2 |
Klhl24
|
kelch-like 24 |
| chr17_+_25059079 | 5.26 |
ENSMUST00000164251.8
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr19_-_34856853 | 5.21 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr7_-_102607982 | 5.20 |
ENSMUST00000216420.2
ENSMUST00000213477.2 |
Olfr575
|
olfactory receptor 575 |
| chr17_+_79922329 | 5.19 |
ENSMUST00000040368.3
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr5_-_87716882 | 5.17 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr4_-_96552349 | 5.15 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr2_+_4722956 | 5.15 |
ENSMUST00000056914.7
|
Bend7
|
BEN domain containing 7 |
| chr1_+_106099482 | 5.14 |
ENSMUST00000061047.7
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chrX_-_7547273 | 5.13 |
ENSMUST00000115695.4
|
Magix
|
MAGI family member, X-linked |
| chr14_-_31362909 | 5.10 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr8_-_64659004 | 5.02 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr14_+_51333816 | 5.01 |
ENSMUST00000169895.3
|
Rnase4
|
ribonuclease, RNase A family 4 |
| chr7_-_133384449 | 4.95 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr4_-_6275629 | 4.90 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr14_-_31362835 | 4.85 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr1_+_171052623 | 4.85 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr10_-_53952686 | 4.82 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr15_+_54975713 | 4.82 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr10_+_87695117 | 4.78 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr19_+_5927821 | 4.76 |
ENSMUST00000145200.8
ENSMUST00000025732.14 ENSMUST00000125114.8 ENSMUST00000155697.8 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr1_+_67162176 | 4.74 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr4_-_104733580 | 4.69 |
ENSMUST00000064873.9
ENSMUST00000106808.10 ENSMUST00000048947.15 |
C8a
|
complement component 8, alpha polypeptide |
| chr5_+_139375540 | 4.66 |
ENSMUST00000100514.3
|
Gpr146
|
G protein-coupled receptor 146 |
| chr13_+_4241149 | 4.62 |
ENSMUST00000021634.4
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr7_-_80052491 | 4.60 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr19_+_5927876 | 4.56 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr6_-_141892517 | 4.55 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr1_-_72251466 | 4.54 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr18_-_3280999 | 4.53 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr1_-_139487951 | 4.50 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr11_-_46280336 | 4.48 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr11_+_118913788 | 4.48 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr15_-_96918203 | 4.48 |
ENSMUST00000166223.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr4_+_104623505 | 4.42 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr4_+_144619647 | 4.42 |
ENSMUST00000154208.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr18_-_6135888 | 4.37 |
ENSMUST00000182383.8
ENSMUST00000062584.14 ENSMUST00000077128.13 ENSMUST00000182038.2 ENSMUST00000182213.8 ENSMUST00000182559.8 |
Arhgap12
|
Rho GTPase activating protein 12 |
| chr2_-_91025492 | 4.36 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr5_-_87572060 | 4.30 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr12_-_84447702 | 4.29 |
ENSMUST00000122194.8
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr4_+_144619696 | 4.28 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr11_-_46280298 | 4.28 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr14_+_55797443 | 4.27 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr19_+_23118545 | 4.21 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9 |
| chr17_+_37253802 | 4.20 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr18_-_60881679 | 4.16 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr11_-_113599778 | 4.16 |
ENSMUST00000106617.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr3_-_146302343 | 4.13 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr19_+_12673147 | 4.11 |
ENSMUST00000025598.10
ENSMUST00000138545.8 ENSMUST00000154822.2 |
Keg1
|
kidney expressed gene 1 |
| chr3_+_137983250 | 4.03 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr11_-_46280281 | 4.00 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr11_+_3981769 | 4.00 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr9_-_45866264 | 3.99 |
ENSMUST00000114573.9
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr6_-_52617288 | 3.97 |
ENSMUST00000031788.9
|
Hibadh
|
3-hydroxyisobutyrate dehydrogenase |
| chr2_+_28531239 | 3.95 |
ENSMUST00000028155.12
ENSMUST00000113869.8 ENSMUST00000113867.9 |
Tsc1
|
TSC complex subunit 1 |
| chr7_-_25176959 | 3.91 |
ENSMUST00000098668.3
ENSMUST00000206687.2 ENSMUST00000206676.2 ENSMUST00000205308.2 ENSMUST00000098669.8 ENSMUST00000206171.2 ENSMUST00000098666.9 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr10_+_87694924 | 3.90 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr14_+_14210932 | 3.86 |
ENSMUST00000022271.14
|
Acox2
|
acyl-Coenzyme A oxidase 2, branched chain |
| chr2_+_34764408 | 3.86 |
ENSMUST00000113068.9
ENSMUST00000047447.13 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr17_+_25097199 | 3.86 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr19_+_42078859 | 3.85 |
ENSMUST00000235932.2
ENSMUST00000066778.6 |
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr12_-_84447625 | 3.80 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr18_-_39622829 | 3.80 |
ENSMUST00000025300.13
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr17_-_31852128 | 3.78 |
ENSMUST00000236909.2
|
Cbs
|
cystathionine beta-synthase |
| chr9_+_114560235 | 3.77 |
ENSMUST00000035007.10
|
Cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr18_-_39622932 | 3.76 |
ENSMUST00000152853.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr14_-_45715308 | 3.72 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr6_+_14901343 | 3.70 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr11_+_70104929 | 3.68 |
ENSMUST00000094055.10
ENSMUST00000126296.8 ENSMUST00000136328.2 ENSMUST00000153993.3 |
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr5_+_90708962 | 3.67 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr12_-_83643883 | 3.67 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr6_-_124888643 | 3.64 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr12_-_83643964 | 3.61 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr1_-_133849131 | 3.59 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr1_+_167445815 | 3.53 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr6_-_138056914 | 3.52 |
ENSMUST00000171804.4
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr3_+_90161470 | 3.51 |
ENSMUST00000029545.15
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr18_-_39622295 | 3.50 |
ENSMUST00000131885.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr15_+_7170004 | 3.48 |
ENSMUST00000226471.2
ENSMUST00000227727.2 |
Lifr
|
LIF receptor alpha |
| chr18_+_45402018 | 3.48 |
ENSMUST00000183850.8
ENSMUST00000066890.14 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr9_+_46179899 | 3.47 |
ENSMUST00000121598.8
|
Apoa5
|
apolipoprotein A-V |
| chr9_+_50405817 | 3.43 |
ENSMUST00000114474.8
ENSMUST00000188047.2 |
Plet1
|
placenta expressed transcript 1 |
| chr15_+_25940912 | 3.42 |
ENSMUST00000226438.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr5_-_45607463 | 3.41 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr2_-_134396268 | 3.41 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr1_+_52158693 | 3.38 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr12_-_71183371 | 3.36 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chrM_+_3906 | 3.36 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr10_-_67972401 | 3.35 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr17_+_31652073 | 3.35 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr19_-_38113696 | 3.35 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr15_-_34495329 | 3.33 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr17_+_31652029 | 3.31 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr15_-_98816012 | 3.30 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr10_-_41487315 | 3.29 |
ENSMUST00000219054.2
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr4_+_138181616 | 3.29 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr3_+_62245765 | 3.27 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr1_+_34275665 | 3.26 |
ENSMUST00000194192.3
|
Dst
|
dystonin |
| chr9_+_44966464 | 3.25 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr1_+_52158721 | 3.23 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr5_-_113285852 | 3.22 |
ENSMUST00000212276.2
|
2900026A02Rik
|
RIKEN cDNA 2900026A02 gene |
| chr15_-_60793115 | 3.22 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr8_+_46111703 | 3.22 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_-_86698484 | 3.22 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr11_+_96920956 | 3.20 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr16_+_22769822 | 3.20 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr2_+_34764496 | 3.18 |
ENSMUST00000028228.6
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr19_-_7780025 | 3.17 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr15_+_25940781 | 3.15 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr2_-_73722932 | 3.15 |
ENSMUST00000154456.8
ENSMUST00000090802.11 ENSMUST00000055833.12 ENSMUST00000112007.8 ENSMUST00000112016.9 |
Atf2
|
activating transcription factor 2 |
| chr2_+_19662529 | 3.12 |
ENSMUST00000052168.6
|
Otud1
|
OTU domain containing 1 |
| chr12_-_104120105 | 3.11 |
ENSMUST00000085050.4
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr6_+_129510145 | 3.10 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr6_+_14901439 | 3.10 |
ENSMUST00000128567.8
|
Foxp2
|
forkhead box P2 |
| chr13_-_63712140 | 3.09 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1 |
| chr8_-_96161414 | 3.08 |
ENSMUST00000211908.2
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr16_+_22769844 | 3.08 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 34.8 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 5.5 | 22.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 5.0 | 24.9 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 4.6 | 13.9 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 3.7 | 15.0 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 3.0 | 21.3 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 2.4 | 7.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 2.3 | 16.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 2.1 | 6.3 | GO:0097037 | heme export(GO:0097037) |
| 2.1 | 8.3 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 2.0 | 6.0 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 2.0 | 10.0 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 2.0 | 7.9 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 2.0 | 7.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 2.0 | 5.9 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.9 | 5.8 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.8 | 7.2 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 1.8 | 10.8 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.8 | 8.8 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 1.7 | 12.0 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.7 | 5.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 1.7 | 17.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.5 | 11.7 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 1.4 | 13.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.4 | 5.7 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 1.3 | 3.9 | GO:0090650 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 1.3 | 5.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 1.3 | 3.9 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 1.3 | 3.8 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 1.2 | 1.2 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 1.2 | 3.5 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 1.2 | 10.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.2 | 4.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 1.1 | 7.9 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 1.1 | 18.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.1 | 4.4 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.0 | 4.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.0 | 6.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.0 | 3.0 | GO:1904735 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 1.0 | 4.9 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 1.0 | 6.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.0 | 7.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.9 | 4.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.9 | 2.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.9 | 5.5 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.9 | 2.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.9 | 2.7 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.9 | 3.6 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.9 | 2.6 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 0.8 | 7.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.8 | 8.5 | GO:0006956 | complement activation(GO:0006956) |
| 0.8 | 4.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.8 | 2.5 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.8 | 5.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.8 | 1.6 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.8 | 23.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.8 | 15.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.8 | 17.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.8 | 3.8 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.8 | 2.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.7 | 5.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.7 | 2.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.7 | 7.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.7 | 18.1 | GO:1901663 | quinone biosynthetic process(GO:1901663) |
| 0.7 | 23.7 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.7 | 2.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.7 | 6.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.7 | 4.7 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.7 | 12.8 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.7 | 2.0 | GO:1901254 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.6 | 1.9 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.6 | 4.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.6 | 7.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.6 | 1.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.6 | 1.9 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.6 | 2.4 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.6 | 1.8 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.6 | 4.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.6 | 12.0 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.6 | 3.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.6 | 13.5 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.6 | 2.9 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.6 | 3.5 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.6 | 6.4 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.6 | 1.7 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.6 | 2.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.6 | 3.9 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.5 | 29.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.5 | 2.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.5 | 5.3 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.5 | 7.8 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 | 2.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.5 | 9.5 | GO:0034030 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.5 | 4.0 | GO:0070268 | cornification(GO:0070268) |
| 0.5 | 3.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.5 | 1.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.5 | 1.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.5 | 1.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.5 | 1.5 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.5 | 12.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.5 | 1.9 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.5 | 2.9 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.5 | 11.0 | GO:0042640 | anagen(GO:0042640) |
| 0.5 | 4.8 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.5 | 1.0 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.5 | 2.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.5 | 1.9 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.5 | 2.7 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.5 | 0.9 | GO:0072240 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.5 | 10.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.5 | 2.7 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.5 | 3.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.4 | 1.8 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.4 | 0.9 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.4 | 1.8 | GO:0002884 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) negative regulation of hypersensitivity(GO:0002884) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.4 | 2.6 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.4 | 5.1 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.4 | 0.8 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.4 | 3.3 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.4 | 1.6 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.4 | 8.9 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.4 | 2.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.4 | 1.6 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 3.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.4 | 1.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.4 | 1.2 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 4.9 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.4 | 8.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 7.8 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.4 | 2.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 1.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.4 | 3.6 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.4 | 0.7 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.3 | 2.8 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 2.4 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.3 | 1.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 3.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.3 | 2.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 1.0 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.3 | 1.0 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.3 | 1.3 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.3 | 1.3 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.3 | 2.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.3 | 1.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 2.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 2.6 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.3 | 1.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 1.6 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 1.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.3 | 3.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.3 | 3.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 7.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.3 | 1.9 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 2.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 2.5 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 | 1.2 | GO:0000239 | pachytene(GO:0000239) RNA 5'-end processing(GO:0000966) |
| 0.3 | 1.8 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 | 1.8 | GO:2000124 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 0.9 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.3 | 1.8 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.3 | 0.9 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.3 | 0.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.3 | 5.5 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 2.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.3 | 0.8 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.3 | 2.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.3 | 0.8 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.3 | 1.7 | GO:0090158 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 1.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.3 | 3.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 2.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 5.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 0.5 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 1.9 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.3 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.3 | 1.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 1.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.3 | 4.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 0.8 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.3 | 2.8 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.3 | 5.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.3 | 0.5 | GO:0032618 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.2 | 3.0 | GO:0042759 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 14.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.2 | 0.7 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.2 | 2.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 1.2 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.2 | 2.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 0.7 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.2 | 1.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.7 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.2 | 2.9 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 3.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 2.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 | 0.2 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.2 | 1.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 0.9 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 1.4 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 1.8 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 4.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 32.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.7 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.2 | 2.0 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.2 | 1.4 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.2 | 4.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 4.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 4.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.2 | 5.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.9 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 1.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.2 | 2.4 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 1.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 1.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 0.8 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.2 | 2.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 0.6 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.2 | 3.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.8 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.2 | 10.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 5.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 0.6 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 0.4 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 | 0.6 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 2.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.2 | GO:0060197 | cloacal septation(GO:0060197) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.2 | 2.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 4.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 2.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.2 | 0.8 | GO:1902045 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.2 | 5.4 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.2 | 3.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 1.0 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 1.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 1.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 1.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.2 | 0.9 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 0.7 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 1.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 0.9 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 1.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 1.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 3.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 0.4 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.2 | 1.0 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.2 | 2.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 0.5 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.2 | 1.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.2 | 2.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 0.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.2 | 1.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.2 | 0.8 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 1.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 2.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 0.6 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 0.5 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 2.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.2 | 0.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 1.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 | 0.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 1.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 2.5 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.2 | 1.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.9 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 0.5 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 0.9 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.4 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.7 | GO:0030576 | Cajal body organization(GO:0030576) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.1 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 2.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.6 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.1 | 1.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 4.5 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 3.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.8 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 6.2 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.1 | 4.7 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 4.7 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 3.3 | GO:0070229 | negative regulation of lymphocyte apoptotic process(GO:0070229) |
| 0.1 | 0.4 | GO:0097273 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.1 | 0.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 3.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 16.1 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.1 | 4.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.8 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 2.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.4 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.9 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.8 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.4 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.3 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 3.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.1 | 0.6 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.9 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.4 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.4 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 | 4.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 1.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.5 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.4 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.1 | 0.5 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 2.9 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 6.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.5 | GO:1903070 | positive regulation of protein lipidation(GO:1903061) negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 1.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.7 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.6 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 1.3 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.1 | 0.3 | GO:0006498 | N-terminal protein lipidation(GO:0006498) N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 1.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 6.8 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.1 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.9 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 1.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 1.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.1 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 8.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.5 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.1 | 0.8 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 2.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.6 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.4 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.1 | 2.2 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.4 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 9.3 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 0.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.7 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.1 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 1.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 1.8 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.1 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.5 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 2.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.3 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.1 | 1.8 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 1.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.5 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.1 | 4.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.4 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.1 | 3.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.6 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 1.5 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.1 | 0.2 | GO:0032700 | negative regulation of interleukin-17 production(GO:0032700) |
| 0.1 | 1.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 1.1 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.2 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 1.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 4.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.5 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 1.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 1.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.5 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 0.4 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.1 | 0.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.7 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.1 | 0.5 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.2 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.5 | GO:0033561 | regulation of water loss via skin(GO:0033561) |
| 0.1 | 2.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 1.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.8 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.7 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.6 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.7 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.1 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.2 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 2.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.2 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.7 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.5 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 1.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 2.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.0 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.0 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 2.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.9 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.2 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 | 0.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 4.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.0 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 1.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.3 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.4 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 0.4 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 1.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.8 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 7.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 1.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.6 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 1.6 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 1.0 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 0.1 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.3 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.5 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.9 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.1 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.4 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 3.0 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.4 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.7 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.3 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.4 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.4 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.9 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.3 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.5 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.6 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.1 | GO:0019236 | response to pheromone(GO:0019236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 25.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 2.3 | 13.5 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 2.1 | 6.3 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.4 | 8.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 1.3 | 14.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.3 | 10.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.1 | 3.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 1.0 | 39.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 1.0 | 3.9 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.9 | 13.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.8 | 2.4 | GO:0044753 | amphisome(GO:0044753) |
| 0.6 | 5.0 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 3.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.6 | 2.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.5 | 2.0 | GO:0019034 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.5 | 2.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 3.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 10.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 1.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.4 | 2.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.4 | 3.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 1.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.4 | 1.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.3 | 3.8 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 5.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 3.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 3.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 2.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 4.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 4.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.3 | 2.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.2 | 0.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 2.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 2.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 6.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 0.7 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 0.7 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 6.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 5.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 11.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 1.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 2.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 0.7 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 4.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 4.3 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.2 | 1.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 12.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 3.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 5.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 0.7 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 40.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 4.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 2.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 3.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 0.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 3.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.4 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.1 | 7.3 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 1.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 12.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 1.5 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.1 | 1.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 6.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 3.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.3 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 9.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 50.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 2.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 6.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.5 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 2.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 3.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.3 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 14.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 14.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 1.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 6.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 6.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.3 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 12.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 7.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 77.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 4.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 5.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 12.0 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 13.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 2.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 29.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 4.7 | 14.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 4.2 | 24.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 3.9 | 11.7 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 3.5 | 13.9 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 2.6 | 10.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 2.5 | 14.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 2.0 | 14.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 2.0 | 6.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.0 | 5.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.9 | 5.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 1.9 | 5.6 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 1.8 | 11.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 1.7 | 13.4 | GO:0005534 | galactose binding(GO:0005534) |
| 1.6 | 8.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 1.6 | 9.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 1.6 | 6.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 1.6 | 4.7 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.6 | 4.7 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 1.4 | 8.7 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.4 | 7.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 1.4 | 10.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.4 | 5.5 | GO:0005118 | sevenless binding(GO:0005118) |
| 1.3 | 4.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.3 | 3.9 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 1.3 | 5.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.3 | 10.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.3 | 28.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.3 | 3.8 | GO:0070026 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitric oxide binding(GO:0070026) |
| 1.3 | 8.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 1.2 | 4.8 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.2 | 12.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 1.2 | 7.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.1 | 3.4 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 1.1 | 3.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 1.0 | 2.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 1.0 | 4.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.0 | 10.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.9 | 2.8 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.9 | 2.8 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.9 | 9.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.9 | 5.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.9 | 3.6 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.8 | 7.6 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.8 | 10.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.8 | 4.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.8 | 4.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.8 | 5.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.8 | 3.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.7 | 8.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.7 | 4.4 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.7 | 3.5 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.7 | 4.7 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.7 | 2.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.7 | 2.6 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.7 | 27.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.7 | 3.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.7 | 5.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.7 | 2.6 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.6 | 7.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.6 | 6.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.6 | 1.9 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.6 | 3.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 19.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.6 | 10.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 2.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.5 | 3.8 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.5 | 1.6 | GO:0070905 | serine binding(GO:0070905) |
| 0.5 | 23.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.5 | 5.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 2.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.5 | 1.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.5 | 2.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 2.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 1.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.5 | 3.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.5 | 2.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.5 | 8.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 9.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 1.8 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.4 | 5.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 2.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.4 | 2.2 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 2.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 1.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.4 | 0.8 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 1.2 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.4 | 4.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 1.6 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.4 | 2.8 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.4 | 2.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 1.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.4 | 3.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 3.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.4 | 2.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 1.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.4 | 4.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.4 | 3.3 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.4 | 2.6 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.4 | 1.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 14.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 6.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 2.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.3 | 1.0 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.3 | 1.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 1.0 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.3 | 4.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 12.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 9.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 18.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.3 | 1.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 6.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 1.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 10.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 8.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 8.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 1.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.3 | 4.1 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.3 | 4.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.3 | 1.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.3 | 2.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 6.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 1.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 1.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 0.8 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 3.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 1.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.7 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 3.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 2.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.2 | 2.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.2 | 3.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 0.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 0.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 1.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 0.9 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.2 | 2.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.7 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.2 | 0.4 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.2 | 2.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 2.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 9.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.2 | 31.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 3.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 2.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.9 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 2.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 1.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 5.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 0.6 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 5.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 1.9 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 1.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 2.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 2.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 2.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 14.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 1.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 2.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 1.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 4.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.4 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 0.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.3 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.1 | 4.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.4 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 4.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 1.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.6 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 0.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 2.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 14.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 2.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 2.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.7 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 4.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 3.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 2.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 6.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 2.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 10.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.5 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 1.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 2.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 3.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 4.9 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 6.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 2.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 2.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.5 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.1 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.1 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 12.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 6.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 2.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.1 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.1 | 4.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 1.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 9.1 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 2.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 3.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 2.0 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 3.4 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.9 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 6.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.5 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 1.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) dynein light chain binding(GO:0045503) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.9 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.4 | 23.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 8.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 23.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 19.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 20.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.3 | 7.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 14.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 10.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 17.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 4.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 7.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 6.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 2.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 4.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 8.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 5.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 4.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 2.1 | 28.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.9 | 15.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.8 | 25.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.5 | 16.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.9 | 19.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.9 | 19.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 3.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.7 | 14.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.6 | 8.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.6 | 1.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.6 | 9.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.5 | 11.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.5 | 12.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 7.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 8.7 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.4 | 1.2 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.4 | 7.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 8.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 38.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 5.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 7.7 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.3 | 13.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 4.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 8.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 3.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 5.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 3.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 6.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 7.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 2.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 6.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 1.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.2 | 3.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 6.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 2.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 2.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 5.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 4.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 3.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 3.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 7.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 20.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 4.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.8 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 5.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 6.3 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 2.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.8 | REACTOME AXON GUIDANCE | Genes involved in Axon guidance |
| 0.1 | 2.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 7.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 0.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | Genes involved in Phospholipase C-mediated cascade |
| 0.0 | 3.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.2 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |