Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Fubp1
Z-value: 0.85
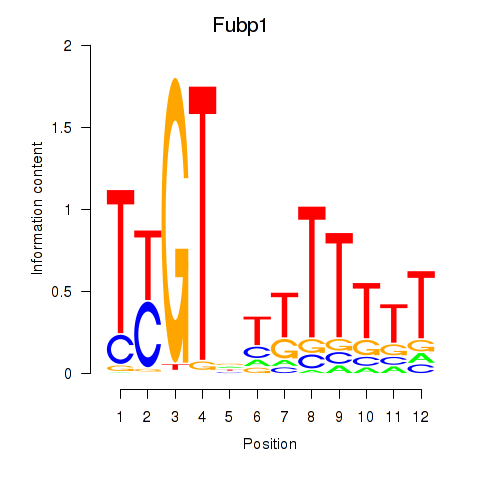
Transcription factors associated with Fubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fubp1
|
ENSMUSG00000028034.16 | Fubp1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fubp1 | mm39_v1_chr3_+_151916059_151916102 | 0.40 | 1.6e-02 | Click! |
Activity profile of Fubp1 motif
Sorted Z-values of Fubp1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Fubp1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60222580 | 3.49 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr4_-_60377932 | 3.25 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr4_-_61259997 | 3.16 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr2_+_103799873 | 1.78 |
ENSMUST00000123437.8
|
Lmo2
|
LIM domain only 2 |
| chr2_+_103800459 | 1.17 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr2_+_103800553 | 1.13 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr3_+_89366425 | 0.83 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr10_+_53472853 | 0.82 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr9_-_48391838 | 0.76 |
ENSMUST00000216470.2
ENSMUST00000217037.2 ENSMUST00000034524.5 ENSMUST00000213895.2 |
Rexo2
|
RNA exonuclease 2 |
| chr11_-_68864666 | 0.73 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr5_+_110987839 | 0.72 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr15_-_57982705 | 0.71 |
ENSMUST00000228783.2
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr10_+_53473032 | 0.71 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr2_-_4656870 | 0.61 |
ENSMUST00000192470.2
ENSMUST00000035721.14 ENSMUST00000152362.7 |
Prpf18
|
pre-mRNA processing factor 18 |
| chr1_+_156193607 | 0.61 |
ENSMUST00000102782.4
|
Gm2000
|
predicted gene 2000 |
| chr11_+_67167950 | 0.59 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr19_-_4384029 | 0.57 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr3_+_89043440 | 0.54 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr11_-_17161504 | 0.52 |
ENSMUST00000020317.8
|
Pno1
|
partner of NOB1 homolog |
| chr5_-_110987604 | 0.51 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr13_+_108452866 | 0.49 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B |
| chrX_-_50106844 | 0.48 |
ENSMUST00000053593.8
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr5_-_146157711 | 0.43 |
ENSMUST00000169407.9
ENSMUST00000161331.8 ENSMUST00000159074.3 ENSMUST00000067837.10 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr13_+_119599287 | 0.43 |
ENSMUST00000026519.10
ENSMUST00000225186.2 ENSMUST00000224081.2 ENSMUST00000224312.2 |
4833420G17Rik
|
RIKEN cDNA 4833420G17 gene |
| chr13_-_46881388 | 0.43 |
ENSMUST00000021803.10
|
Nup153
|
nucleoporin 153 |
| chr9_+_106699073 | 0.41 |
ENSMUST00000159645.8
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr4_+_109200225 | 0.40 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr6_+_134897364 | 0.39 |
ENSMUST00000067327.11
ENSMUST00000003115.9 |
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr13_-_21964747 | 0.38 |
ENSMUST00000080511.3
|
H1f5
|
H1.5 linker histone, cluster member |
| chr5_+_107655487 | 0.38 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr2_-_103609703 | 0.37 |
ENSMUST00000143188.2
|
Caprin1
|
cell cycle associated protein 1 |
| chr11_-_40586029 | 0.36 |
ENSMUST00000101347.10
|
Mat2b
|
methionine adenosyltransferase II, beta |
| chr13_+_108452930 | 0.36 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B |
| chr2_+_103900164 | 0.36 |
ENSMUST00000111131.9
ENSMUST00000111132.8 ENSMUST00000129749.8 |
Cd59b
|
CD59b antigen |
| chr7_-_110462446 | 0.34 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr11_-_86248395 | 0.34 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr3_+_122717852 | 0.34 |
ENSMUST00000106429.6
|
1810037I17Rik
|
RIKEN cDNA 1810037I17 gene |
| chr8_-_57940834 | 0.33 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide |
| chr10_+_94412116 | 0.33 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr13_-_3943433 | 0.32 |
ENSMUST00000222504.2
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chr14_-_72840373 | 0.32 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr1_-_36982747 | 0.32 |
ENSMUST00000185964.3
|
Tmem131
|
transmembrane protein 131 |
| chr18_-_43610829 | 0.31 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr17_-_46343291 | 0.30 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr7_+_101619897 | 0.30 |
ENSMUST00000211272.2
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr2_+_90817948 | 0.29 |
ENSMUST00000111452.8
ENSMUST00000111455.9 |
Celf1
|
CUGBP, Elav-like family member 1 |
| chr9_-_103242096 | 0.29 |
ENSMUST00000116517.9
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr8_-_61407760 | 0.28 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr11_-_78074377 | 0.28 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr12_-_32258331 | 0.28 |
ENSMUST00000220366.2
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr2_-_93988229 | 0.27 |
ENSMUST00000028619.5
|
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr18_-_34884555 | 0.27 |
ENSMUST00000060710.9
|
Cdc25c
|
cell division cycle 25C |
| chr5_-_135423353 | 0.26 |
ENSMUST00000111171.6
|
Pom121
|
nuclear pore membrane protein 121 |
| chr6_+_134897473 | 0.26 |
ENSMUST00000204807.2
|
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr2_+_173918715 | 0.26 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr6_+_35154545 | 0.25 |
ENSMUST00000170234.2
|
Nup205
|
nucleoporin 205 |
| chr3_+_84859453 | 0.25 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr16_-_92622972 | 0.25 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr1_-_138103021 | 0.24 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr16_+_17093941 | 0.24 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr19_+_41471395 | 0.24 |
ENSMUST00000237208.2
ENSMUST00000238398.2 |
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr5_-_33011530 | 0.24 |
ENSMUST00000130134.3
ENSMUST00000120129.9 |
Prr14l
|
proline rich 14-like |
| chr3_-_100591906 | 0.24 |
ENSMUST00000130066.2
|
Man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr12_-_32258469 | 0.24 |
ENSMUST00000085469.6
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr12_-_32258604 | 0.23 |
ENSMUST00000053215.14
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr11_+_54517164 | 0.23 |
ENSMUST00000239168.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr6_+_30401864 | 0.22 |
ENSMUST00000068240.13
ENSMUST00000068259.10 ENSMUST00000132581.8 |
Klhdc10
|
kelch domain containing 10 |
| chr11_+_67090878 | 0.22 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr6_+_83142902 | 0.22 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr4_+_118266526 | 0.22 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr10_+_94350687 | 0.21 |
ENSMUST00000065060.12
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr12_+_71095112 | 0.21 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr13_-_103901010 | 0.21 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr3_+_138233004 | 0.21 |
ENSMUST00000196990.5
ENSMUST00000200239.5 ENSMUST00000200100.2 |
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr11_+_4845328 | 0.21 |
ENSMUST00000038237.8
|
Thoc5
|
THO complex 5 |
| chr10_+_126911134 | 0.20 |
ENSMUST00000239120.2
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr6_-_34887530 | 0.20 |
ENSMUST00000149448.8
ENSMUST00000133336.8 |
Wdr91
|
WD repeat domain 91 |
| chr10_-_53506038 | 0.20 |
ENSMUST00000218549.3
|
Mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr12_+_55431007 | 0.18 |
ENSMUST00000163070.8
|
Psma6
|
proteasome subunit alpha 6 |
| chr9_+_61279640 | 0.17 |
ENSMUST00000178113.8
ENSMUST00000159386.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr19_-_56536443 | 0.17 |
ENSMUST00000182059.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr6_+_29361408 | 0.17 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chrX_+_162694397 | 0.17 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr6_+_48624295 | 0.17 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr15_-_5093222 | 0.17 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr4_+_118266582 | 0.17 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr17_+_26471870 | 0.16 |
ENSMUST00000025023.15
|
Luc7l
|
Luc7-like |
| chr7_-_110681402 | 0.16 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr11_+_11414256 | 0.16 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr1_+_179788675 | 0.16 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr4_-_118266416 | 0.16 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr19_+_53128861 | 0.16 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr17_+_26471889 | 0.16 |
ENSMUST00000114976.9
ENSMUST00000140427.8 ENSMUST00000119928.8 |
Luc7l
|
Luc7-like |
| chr1_-_134883645 | 0.16 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_+_122723365 | 0.15 |
ENSMUST00000205514.2
ENSMUST00000094053.7 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr3_+_85946145 | 0.15 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr13_+_96524802 | 0.15 |
ENSMUST00000099295.6
|
Poc5
|
POC5 centriolar protein |
| chr11_+_4845314 | 0.15 |
ENSMUST00000101615.9
|
Thoc5
|
THO complex 5 |
| chr18_-_70274639 | 0.15 |
ENSMUST00000121693.8
|
Rab27b
|
RAB27B, member RAS oncogene family |
| chr1_-_138102972 | 0.14 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_79974166 | 0.14 |
ENSMUST00000047362.11
ENSMUST00000121882.8 |
Rccd1
|
RCC1 domain containing 1 |
| chr15_-_98851566 | 0.13 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr8_+_66838927 | 0.13 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr1_+_179788037 | 0.13 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_-_45001141 | 0.12 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr11_+_99748741 | 0.12 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr14_+_61836944 | 0.12 |
ENSMUST00000039562.8
|
Trim13
|
tripartite motif-containing 13 |
| chr19_+_44551280 | 0.12 |
ENSMUST00000040455.5
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr3_-_146487102 | 0.12 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr4_-_147726953 | 0.11 |
ENSMUST00000133006.2
ENSMUST00000037565.14 ENSMUST00000105720.8 |
Zfp979
|
zinc finger protein 979 |
| chr9_+_108673171 | 0.11 |
ENSMUST00000195514.6
ENSMUST00000085018.6 ENSMUST00000192028.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr3_-_108934916 | 0.11 |
ENSMUST00000171143.2
|
Fam102b
|
family with sequence similarity 102, member B |
| chr5_-_124492734 | 0.10 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chrX_-_166638057 | 0.10 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr12_+_17316546 | 0.10 |
ENSMUST00000057288.7
ENSMUST00000239402.2 |
Pdia6
|
protein disulfide isomerase associated 6 |
| chr19_-_40982576 | 0.09 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr4_-_94538370 | 0.08 |
ENSMUST00000053419.9
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr5_-_66308666 | 0.08 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr19_-_56536646 | 0.08 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr11_-_99742434 | 0.08 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr10_-_116417333 | 0.07 |
ENSMUST00000218744.2
ENSMUST00000105267.8 ENSMUST00000105265.8 ENSMUST00000167706.8 ENSMUST00000168036.8 ENSMUST00000169921.8 ENSMUST00000020374.6 |
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr19_-_47680528 | 0.07 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr4_-_55532453 | 0.07 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr3_-_151960948 | 0.07 |
ENSMUST00000199423.5
ENSMUST00000198460.5 |
Nexn
|
nexilin |
| chr18_-_77855446 | 0.07 |
ENSMUST00000048192.9
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr13_-_96572460 | 0.06 |
ENSMUST00000181761.2
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr18_-_84104574 | 0.06 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr12_-_87519032 | 0.06 |
ENSMUST00000021428.9
|
Snw1
|
SNW domain containing 1 |
| chr12_-_84031622 | 0.06 |
ENSMUST00000164935.3
|
Heatr4
|
HEAT repeat containing 4 |
| chr10_+_39296005 | 0.05 |
ENSMUST00000157009.8
|
Fyn
|
Fyn proto-oncogene |
| chr13_+_83672965 | 0.05 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chrX_+_93679671 | 0.05 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr5_+_4073343 | 0.05 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr9_-_103079312 | 0.05 |
ENSMUST00000035157.10
|
Srprb
|
signal recognition particle receptor, B subunit |
| chr6_+_17307639 | 0.05 |
ENSMUST00000115453.2
|
Cav1
|
caveolin 1, caveolae protein |
| chrX_-_58613428 | 0.04 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chr3_-_151960992 | 0.04 |
ENSMUST00000198750.5
|
Nexn
|
nexilin |
| chr6_-_113354468 | 0.04 |
ENSMUST00000099118.8
|
Tada3
|
transcriptional adaptor 3 |
| chr2_+_104420798 | 0.04 |
ENSMUST00000028599.8
|
Cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr9_+_88721217 | 0.03 |
ENSMUST00000163255.9
ENSMUST00000186363.2 |
Trim43c
|
tripartite motif-containing 43C |
| chr3_+_96537484 | 0.03 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr18_-_32044877 | 0.03 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr10_-_12689345 | 0.03 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr6_-_99005212 | 0.03 |
ENSMUST00000177437.8
ENSMUST00000177229.8 ENSMUST00000124058.8 |
Foxp1
|
forkhead box P1 |
| chr10_+_127226180 | 0.03 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr7_+_98089623 | 0.03 |
ENSMUST00000206435.2
|
Gucy2d
|
guanylate cyclase 2d |
| chr6_-_25689781 | 0.03 |
ENSMUST00000200812.2
|
Gpr37
|
G protein-coupled receptor 37 |
| chr3_+_154369496 | 0.03 |
ENSMUST00000098496.9
|
Erich3
|
glutamate rich 3 |
| chr1_-_134883577 | 0.02 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr18_-_84104507 | 0.02 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr3_+_96537235 | 0.02 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr16_-_92622659 | 0.02 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chrX_-_161612373 | 0.02 |
ENSMUST00000041370.11
ENSMUST00000112316.9 ENSMUST00000112315.2 |
Txlng
|
taxilin gamma |
| chr6_+_113366207 | 0.02 |
ENSMUST00000204026.3
|
Ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr6_+_18866339 | 0.02 |
ENSMUST00000115396.7
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr17_+_35268942 | 0.01 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr7_-_126165530 | 0.01 |
ENSMUST00000180459.3
ENSMUST00000032992.7 |
Eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr4_-_94538329 | 0.01 |
ENSMUST00000107101.2
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr4_-_130169006 | 0.01 |
ENSMUST00000122374.8
|
Serinc2
|
serine incorporator 2 |
| chr4_-_132649798 | 0.00 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr14_+_51366512 | 0.00 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 0.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 0.6 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.2 | 0.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.4 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 1.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.3 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.1 | 0.4 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.7 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.3 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.3 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.6 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.1 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.4 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.6 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.5 | GO:0090557 | Rap protein signal transduction(GO:0032486) establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.3 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.2 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 4.1 | GO:0070888 | bHLH transcription factor binding(GO:0043425) E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.8 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |