Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
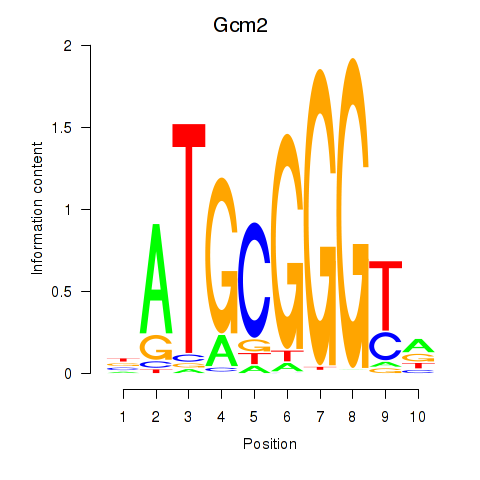
Results for Gcm2
Z-value: 0.86
Transcription factors associated with Gcm2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gcm2
|
ENSMUSG00000021362.8 | Gcm2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gcm2 | mm39_v1_chr13_-_41264511_41264511 | 0.17 | 3.3e-01 | Click! |
Activity profile of Gcm2 motif
Sorted Z-values of Gcm2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gcm2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_33136021 | 3.97 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr17_-_33136277 | 3.64 |
ENSMUST00000234538.2
ENSMUST00000235058.2 ENSMUST00000234759.2 ENSMUST00000179434.8 ENSMUST00000234797.2 |
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr15_+_76579960 | 2.23 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr15_+_76579885 | 1.93 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr1_-_180021039 | 1.86 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr14_+_66208253 | 1.72 |
ENSMUST00000138191.8
|
Clu
|
clusterin |
| chr2_+_155359868 | 1.64 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr11_+_98239230 | 1.62 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr10_+_128089965 | 1.61 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr7_-_34914675 | 1.59 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr2_+_155360015 | 1.50 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr8_-_71990085 | 1.47 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr7_-_105249308 | 1.43 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr7_-_99276310 | 1.35 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr14_+_66208498 | 1.34 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr17_-_24915037 | 1.33 |
ENSMUST00000234235.2
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr7_-_140590605 | 1.30 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr2_-_91466739 | 1.28 |
ENSMUST00000111335.2
ENSMUST00000028681.15 |
F2
|
coagulation factor II |
| chr11_-_113600346 | 1.26 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr5_-_108823435 | 1.26 |
ENSMUST00000051757.14
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr7_-_13571334 | 1.25 |
ENSMUST00000108522.5
|
Sult2a1
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr8_-_118400418 | 1.23 |
ENSMUST00000173522.8
ENSMUST00000174450.2 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr10_+_93324624 | 1.13 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase |
| chr7_+_29883569 | 1.12 |
ENSMUST00000098594.4
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr14_+_66208613 | 1.11 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr7_-_127534601 | 1.10 |
ENSMUST00000141385.7
ENSMUST00000156152.3 |
Prss36
|
protease, serine 36 |
| chr5_+_146016064 | 1.09 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr16_+_22769822 | 1.06 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chrX_+_149377416 | 1.05 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_-_60793115 | 1.02 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr17_-_34219225 | 1.01 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr5_-_123313978 | 0.99 |
ENSMUST00000144679.2
|
Hpd
|
4-hydroxyphenylpyruvic acid dioxygenase |
| chr7_+_30676465 | 0.98 |
ENSMUST00000058093.6
|
Fam187b
|
family with sequence similarity 187, member B |
| chr17_+_35481702 | 0.96 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr17_+_35482063 | 0.94 |
ENSMUST00000172503.3
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr1_-_192923816 | 0.93 |
ENSMUST00000160929.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr17_-_57366795 | 0.93 |
ENSMUST00000040280.14
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr9_-_108444561 | 0.92 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr8_-_70355208 | 0.90 |
ENSMUST00000110167.5
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr1_+_182591771 | 0.89 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr16_+_22769844 | 0.89 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr11_+_77928736 | 0.89 |
ENSMUST00000072289.12
ENSMUST00000100784.9 ENSMUST00000073660.7 |
Flot2
|
flotillin 2 |
| chr17_+_32904629 | 0.88 |
ENSMUST00000008801.7
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr11_-_113600838 | 0.88 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr9_-_21900724 | 0.87 |
ENSMUST00000045726.8
|
Rgl3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr16_-_38342949 | 0.86 |
ENSMUST00000002925.6
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr7_-_46365108 | 0.86 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr11_+_69826603 | 0.86 |
ENSMUST00000018698.12
|
Ybx2
|
Y box protein 2 |
| chr9_-_21913833 | 0.85 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr2_-_32341408 | 0.85 |
ENSMUST00000028160.15
ENSMUST00000113310.9 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr3_+_97536120 | 0.85 |
ENSMUST00000107050.8
ENSMUST00000029729.15 ENSMUST00000107049.2 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr7_+_29883611 | 0.84 |
ENSMUST00000208441.2
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr2_+_70305267 | 0.83 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr5_-_77262968 | 0.82 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr5_-_73495895 | 0.82 |
ENSMUST00000202012.2
|
Ociad2
|
OCIA domain containing 2 |
| chr7_-_12732067 | 0.82 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr17_+_35780977 | 0.82 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr13_+_49340995 | 0.82 |
ENSMUST00000120733.8
|
Ninj1
|
ninjurin 1 |
| chr14_+_40827108 | 0.81 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr6_+_125298372 | 0.80 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr17_+_24955647 | 0.79 |
ENSMUST00000101800.7
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr8_-_45786226 | 0.79 |
ENSMUST00000095328.6
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr19_+_8815024 | 0.79 |
ENSMUST00000159634.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr16_-_84632383 | 0.79 |
ENSMUST00000114191.8
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr9_+_46179899 | 0.78 |
ENSMUST00000121598.8
|
Apoa5
|
apolipoprotein A-V |
| chr17_-_84154173 | 0.78 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr11_-_120551426 | 0.77 |
ENSMUST00000106177.8
|
Notum
|
notum palmitoleoyl-protein carboxylesterase |
| chr4_-_155445818 | 0.77 |
ENSMUST00000030922.15
|
Prkcz
|
protein kinase C, zeta |
| chr17_-_24915121 | 0.77 |
ENSMUST00000046839.10
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr4_+_140966810 | 0.77 |
ENSMUST00000141834.9
|
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr8_-_122671588 | 0.77 |
ENSMUST00000057653.8
|
Car5a
|
carbonic anhydrase 5a, mitochondrial |
| chr6_-_86503178 | 0.77 |
ENSMUST00000053015.7
|
Pcbp1
|
poly(rC) binding protein 1 |
| chr15_+_76215488 | 0.77 |
ENSMUST00000172281.8
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr2_+_155078522 | 0.76 |
ENSMUST00000150602.2
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr11_-_120714921 | 0.76 |
ENSMUST00000206589.2
|
Fasn
|
fatty acid synthase |
| chr16_-_17745999 | 0.76 |
ENSMUST00000003622.16
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr4_+_138032035 | 0.76 |
ENSMUST00000030538.5
|
Ddost
|
dolichyl-di-phosphooligosaccharide-protein glycotransferase |
| chr15_-_82648376 | 0.76 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr13_+_31809774 | 0.76 |
ENSMUST00000042054.3
|
Foxf2
|
forkhead box F2 |
| chr8_+_105996469 | 0.74 |
ENSMUST00000172525.8
ENSMUST00000174837.8 ENSMUST00000173859.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr7_-_84254973 | 0.74 |
ENSMUST00000032865.17
|
Fah
|
fumarylacetoacetate hydrolase |
| chr7_-_12731594 | 0.74 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr7_+_100966289 | 0.73 |
ENSMUST00000163799.9
ENSMUST00000164479.9 |
Stard10
|
START domain containing 10 |
| chr12_-_103956176 | 0.72 |
ENSMUST00000151709.3
ENSMUST00000176246.3 ENSMUST00000074693.13 ENSMUST00000120251.9 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr2_+_164404499 | 0.72 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr16_-_84632439 | 0.72 |
ENSMUST00000138279.2
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr7_+_29931309 | 0.71 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr4_+_129229373 | 0.70 |
ENSMUST00000141235.8
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr12_-_103623418 | 0.70 |
ENSMUST00000044159.7
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr1_-_121255448 | 0.70 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr4_+_106924181 | 0.68 |
ENSMUST00000106758.8
ENSMUST00000145324.8 ENSMUST00000106760.8 |
Cyb5rl
|
cytochrome b5 reductase-like |
| chr9_-_121745354 | 0.68 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr15_-_76191301 | 0.67 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr17_+_34043536 | 0.67 |
ENSMUST00000048249.8
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr7_-_48497771 | 0.67 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr4_+_138181616 | 0.67 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr17_+_24955613 | 0.66 |
ENSMUST00000115262.9
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr19_-_6117815 | 0.66 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr19_-_4489415 | 0.66 |
ENSMUST00000235680.2
ENSMUST00000117462.2 ENSMUST00000048197.10 |
Rhod
|
ras homolog family member D |
| chr10_-_81262948 | 0.66 |
ENSMUST00000078185.14
ENSMUST00000020461.15 ENSMUST00000105321.10 |
Nfic
|
nuclear factor I/C |
| chr10_-_75633362 | 0.66 |
ENSMUST00000120177.8
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr1_+_171238911 | 0.64 |
ENSMUST00000160486.8
|
Usf1
|
upstream transcription factor 1 |
| chr3_-_113166153 | 0.64 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr11_+_70104736 | 0.63 |
ENSMUST00000171032.8
|
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr6_+_54016543 | 0.63 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr4_-_116991150 | 0.62 |
ENSMUST00000076859.12
|
Plk3
|
polo like kinase 3 |
| chr8_+_105996419 | 0.62 |
ENSMUST00000036127.9
ENSMUST00000163734.9 |
Hsf4
|
heat shock transcription factor 4 |
| chr2_-_91025441 | 0.62 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr2_+_122607297 | 0.62 |
ENSMUST00000124460.2
ENSMUST00000147475.2 |
Sqor
|
sulfide quinone oxidoreductase |
| chr7_+_100970435 | 0.62 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr11_-_69586347 | 0.62 |
ENSMUST00000181261.2
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr13_-_42001075 | 0.62 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr11_+_68858942 | 0.62 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr12_-_85871281 | 0.61 |
ENSMUST00000021676.12
ENSMUST00000142331.2 |
Erg28
|
ergosterol biosynthesis 28 |
| chr17_+_46991972 | 0.61 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr5_-_87682972 | 0.61 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr2_+_155078449 | 0.61 |
ENSMUST00000109682.9
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr10_-_80634293 | 0.60 |
ENSMUST00000218209.2
ENSMUST00000036805.7 |
Plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr6_-_23839136 | 0.60 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr14_-_55983142 | 0.59 |
ENSMUST00000002403.10
|
Dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr9_-_108141105 | 0.59 |
ENSMUST00000166905.8
ENSMUST00000191899.6 |
Dag1
|
dystroglycan 1 |
| chr5_-_120641658 | 0.58 |
ENSMUST00000031597.7
|
Plbd2
|
phospholipase B domain containing 2 |
| chr17_-_31855605 | 0.58 |
ENSMUST00000151718.3
ENSMUST00000135425.9 ENSMUST00000155814.8 |
Cbs
|
cystathionine beta-synthase |
| chr1_+_132226308 | 0.58 |
ENSMUST00000046071.5
|
Klhdc8a
|
kelch domain containing 8A |
| chr6_+_86342622 | 0.58 |
ENSMUST00000071492.9
|
Fam136a
|
family with sequence similarity 136, member A |
| chr11_+_70410445 | 0.57 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr2_-_91025380 | 0.57 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_+_88093726 | 0.57 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr11_-_62172164 | 0.56 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr11_-_73067828 | 0.56 |
ENSMUST00000108480.2
ENSMUST00000054952.4 |
Emc6
|
ER membrane protein complex subunit 6 |
| chr8_-_29709652 | 0.56 |
ENSMUST00000168630.4
|
Unc5d
|
unc-5 netrin receptor D |
| chr19_+_8641369 | 0.56 |
ENSMUST00000035444.10
ENSMUST00000163785.2 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
| chr4_-_46536088 | 0.56 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr6_+_129510331 | 0.55 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr15_-_100534572 | 0.55 |
ENSMUST00000230280.2
|
Smagp
|
small cell adhesion glycoprotein |
| chr2_-_91025492 | 0.55 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_-_127307898 | 0.55 |
ENSMUST00000207019.2
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr8_+_107877252 | 0.55 |
ENSMUST00000034400.5
|
Cyb5b
|
cytochrome b5 type B |
| chr13_-_42001102 | 0.55 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr2_-_32321116 | 0.55 |
ENSMUST00000127961.3
ENSMUST00000136361.8 ENSMUST00000052119.14 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr7_-_19530714 | 0.54 |
ENSMUST00000108449.9
ENSMUST00000043822.8 |
Cblc
|
Casitas B-lineage lymphoma c |
| chr15_+_31568937 | 0.54 |
ENSMUST00000162532.8
ENSMUST00000070918.14 |
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr14_+_40826970 | 0.54 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr9_-_26910833 | 0.54 |
ENSMUST00000060513.8
ENSMUST00000120367.8 |
Acad8
|
acyl-Coenzyme A dehydrogenase family, member 8 |
| chr4_-_155445779 | 0.54 |
ENSMUST00000105624.2
|
Prkcz
|
protein kinase C, zeta |
| chr15_+_99488527 | 0.54 |
ENSMUST00000169082.3
ENSMUST00000088200.13 |
Aqp5
|
aquaporin 5 |
| chr12_-_102671154 | 0.54 |
ENSMUST00000178697.2
ENSMUST00000046518.12 |
Itpk1
|
inositol 1,3,4-triphosphate 5/6 kinase |
| chr17_+_34138611 | 0.53 |
ENSMUST00000234247.2
|
Tapbp
|
TAP binding protein |
| chr7_-_127308059 | 0.53 |
ENSMUST00000061468.9
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr15_+_100768551 | 0.53 |
ENSMUST00000082209.13
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr5_+_147797258 | 0.53 |
ENSMUST00000031654.10
|
Pomp
|
proteasome maturation protein |
| chr7_+_143383363 | 0.52 |
ENSMUST00000124340.8
|
Dhcr7
|
7-dehydrocholesterol reductase |
| chr5_-_121045568 | 0.52 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr8_-_29709455 | 0.52 |
ENSMUST00000211448.2
ENSMUST00000210298.2 ENSMUST00000209401.2 ENSMUST00000210785.2 |
Unc5d
|
unc-5 netrin receptor D |
| chr11_+_98938137 | 0.52 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr2_-_26823793 | 0.52 |
ENSMUST00000154651.2
ENSMUST00000015011.10 |
Surf4
|
surfeit gene 4 |
| chr15_-_31367668 | 0.52 |
ENSMUST00000110410.10
ENSMUST00000076942.5 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr5_-_24652775 | 0.52 |
ENSMUST00000123167.2
ENSMUST00000030799.15 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr11_-_120552001 | 0.51 |
ENSMUST00000150458.2
|
Notum
|
notum palmitoleoyl-protein carboxylesterase |
| chr6_+_88701810 | 0.51 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr8_+_70536103 | 0.51 |
ENSMUST00000007738.11
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr4_+_108691798 | 0.50 |
ENSMUST00000030296.9
|
Txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr7_-_127307791 | 0.50 |
ENSMUST00000205977.2
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr11_+_115494751 | 0.50 |
ENSMUST00000058109.9
|
Mrps7
|
mitchondrial ribosomal protein S7 |
| chr9_-_103105638 | 0.49 |
ENSMUST00000126359.2
|
Trf
|
transferrin |
| chr14_+_53588485 | 0.49 |
ENSMUST00000199280.2
ENSMUST00000103633.2 |
Trav16n
|
T cell receptor alpha variable 16n |
| chr11_-_120715351 | 0.49 |
ENSMUST00000055655.9
|
Fasn
|
fatty acid synthase |
| chr17_+_46992101 | 0.49 |
ENSMUST00000233612.2
|
Mea1
|
male enhanced antigen 1 |
| chr15_+_25843225 | 0.49 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr11_-_55352005 | 0.48 |
ENSMUST00000108857.2
|
Atox1
|
antioxidant 1 copper chaperone |
| chr2_+_144435974 | 0.48 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr16_-_56987670 | 0.48 |
ENSMUST00000023432.10
|
Nit2
|
nitrilase family, member 2 |
| chr5_+_90708962 | 0.48 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr9_+_45817795 | 0.48 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr16_-_84632513 | 0.48 |
ENSMUST00000023608.14
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr6_+_99669640 | 0.48 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr9_-_106769069 | 0.48 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr15_+_26309125 | 0.48 |
ENSMUST00000126304.2
ENSMUST00000140840.8 ENSMUST00000152841.2 |
Marchf11
|
membrane associated ring-CH-type finger 11 |
| chr5_+_28370687 | 0.48 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr4_-_96441854 | 0.48 |
ENSMUST00000030303.6
|
Cyp2j6
|
cytochrome P450, family 2, subfamily j, polypeptide 6 |
| chr17_-_35340006 | 0.47 |
ENSMUST00000174306.8
ENSMUST00000174024.8 |
Csnk2b
|
casein kinase 2, beta polypeptide |
| chr3_+_89680867 | 0.47 |
ENSMUST00000038356.13
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr13_-_42000958 | 0.47 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr4_-_40279382 | 0.47 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr17_+_25114090 | 0.46 |
ENSMUST00000043907.14
|
Mrps34
|
mitochondrial ribosomal protein S34 |
| chr11_+_94544593 | 0.46 |
ENSMUST00000025278.8
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr15_+_99290832 | 0.46 |
ENSMUST00000160635.8
ENSMUST00000161250.8 ENSMUST00000229392.2 ENSMUST00000161778.8 |
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr9_-_106769131 | 0.46 |
ENSMUST00000159283.8
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr8_+_85786684 | 0.46 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr8_-_106670014 | 0.46 |
ENSMUST00000038896.8
|
Lcat
|
lecithin cholesterol acyltransferase |
| chrX_+_68403900 | 0.46 |
ENSMUST00000033532.7
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr5_+_139529643 | 0.46 |
ENSMUST00000174792.2
|
Uncx
|
UNC homeobox |
| chr7_-_98831916 | 0.46 |
ENSMUST00000033001.6
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr18_-_35788255 | 0.46 |
ENSMUST00000190196.5
|
Prob1
|
proline rich basic protein 1 |
| chr6_-_125357756 | 0.45 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr11_-_66416790 | 0.45 |
ENSMUST00000066679.7
|
Shisa6
|
shisa family member 6 |
| chr6_-_85809064 | 0.45 |
ENSMUST00000032073.7
|
Nat8
|
N-acetyltransferase 8 (GCN5-related) |
| chr12_+_113104085 | 0.45 |
ENSMUST00000200380.5
|
Crip2
|
cysteine rich protein 2 |
| chr11_-_53313950 | 0.45 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr18_-_35760260 | 0.45 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.7 | 4.2 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.6 | 1.9 | GO:0097037 | heme export(GO:0097037) |
| 0.6 | 2.9 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.5 | 1.4 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 0.4 | 1.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.4 | 1.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.4 | 1.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 1.2 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.4 | 1.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.4 | 1.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 1.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.4 | 1.8 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 1.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 0.9 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.3 | 1.5 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 0.8 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.3 | 1.3 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 0.7 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.2 | 0.9 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.2 | 1.1 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.2 | 0.9 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.2 | 0.7 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.2 | 0.9 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.2 | 0.7 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 0.2 | 0.6 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 0.8 | GO:0050822 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 0.6 | GO:0016129 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.2 | 1.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 0.8 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.2 | 0.6 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.2 | 2.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.6 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 1.3 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 0.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.7 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.2 | 0.8 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 2.1 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 0.5 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.2 | 0.5 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.2 | 1.3 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.2 | 0.8 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.2 | 0.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 0.8 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 0.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.2 | 0.5 | GO:0018003 | peptidyl-lysine N6-acetylation(GO:0018003) |
| 0.2 | 0.5 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.9 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.1 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.1 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.1 | 1.3 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.1 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 8.5 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 1.4 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 0.3 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.1 | 0.3 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.1 | 0.3 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.1 | 0.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.4 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.4 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.1 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 0.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.4 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 1.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.5 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.4 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.4 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.3 | GO:1903826 | mitochondrial ornithine transport(GO:0000066) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) L-lysine transmembrane transport(GO:1903401) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 2.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.5 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.6 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.8 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.4 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.3 | GO:2000742 | cervix development(GO:0060067) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.8 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.3 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 1.6 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.3 | GO:1903630 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.5 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 2.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 1.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.2 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.8 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.5 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.4 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.4 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.8 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 0.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.1 | 0.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.6 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.2 | GO:0002660 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 1.6 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 2.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.3 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.1 | 2.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.4 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 1.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 0.2 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 | 0.2 | GO:0061723 | glycophagy(GO:0061723) |
| 0.1 | 0.2 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 | 0.7 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.2 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 0.1 | 0.5 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.4 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.1 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.5 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.4 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0071931 | Cajal body organization(GO:0030576) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0060197 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 0.0 | 0.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 1.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.6 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.4 | GO:0006568 | tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 1.8 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.2 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 1.0 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.2 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.3 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.0 | 0.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 1.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.0 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 1.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.5 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 2.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.7 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.5 | GO:0021889 | olfactory bulb interneuron differentiation(GO:0021889) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 1.1 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.3 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.6 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.2 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.5 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0070779 | sulfur amino acid transport(GO:0000101) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.0 | 0.1 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 0.0 | 0.2 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.0 | 0.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.4 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.2 | GO:0038095 | extracellular matrix-cell signaling(GO:0035426) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0044598 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0090156 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.8 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0035720 | signal transduction downstream of smoothened(GO:0007227) intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:1900193 | negative regulation of oocyte development(GO:0060283) regulation of oocyte maturation(GO:1900193) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.6 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:1904173 | regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.0 | 0.2 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.3 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0021886 | female meiosis I(GO:0007144) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.3 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0060767 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.7 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.0 | 0.2 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 1.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.5 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.0 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.4 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.4 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 1.0 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.3 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.1 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 1.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.0 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.0 | 0.2 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.3 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.6 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.0 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:1902653 | secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.9 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.5 | 1.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 3.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 4.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 0.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.2 | 0.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 1.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 0.7 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 0.8 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 0.5 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 1.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 2.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 1.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 2.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 1.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 2.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 2.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.3 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 4.8 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 3.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 6.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 6.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 5.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.0 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.8 | 4.2 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.6 | 3.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 1.6 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.4 | 1.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.4 | 2.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.4 | 1.3 | GO:0016296 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) phosphopantetheine binding(GO:0031177) |
| 0.3 | 1.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 1.0 | GO:0008127 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) quercetin 2,3-dioxygenase activity(GO:0008127) |
| 0.3 | 0.9 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 1.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 4.8 | GO:0046977 | TAP binding(GO:0046977) |
| 0.3 | 0.8 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.2 | 1.5 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 0.7 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 0.9 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 0.7 | GO:0047651 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.2 | 1.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 2.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 1.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 1.4 | GO:0050656 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 0.8 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 0.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitric oxide binding(GO:0070026) |
| 0.2 | 0.7 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 0.5 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.2 | 0.5 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.2 | 4.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.8 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.2 | 0.8 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 0.5 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.9 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 1.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.7 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.4 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.7 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.4 | GO:0004371 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.1 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.4 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.4 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 0.5 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 1.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 1.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 0.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.3 | GO:0005287 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.1 | 0.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.5 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.4 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 2.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.2 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.1 | 0.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.6 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 0.1 | 2.0 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.3 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.3 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.6 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.3 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.9 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.0 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 1.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.7 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 1.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.6 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 1.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 1.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 2.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.0 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 3.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 4.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.9 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 1.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.2 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 5.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 3.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |