Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Glis2
Z-value: 1.22
Transcription factors associated with Glis2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis2
|
ENSMUSG00000014303.14 | Glis2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis2 | mm39_v1_chr16_+_4412546_4412599 | 0.61 | 6.9e-05 | Click! |
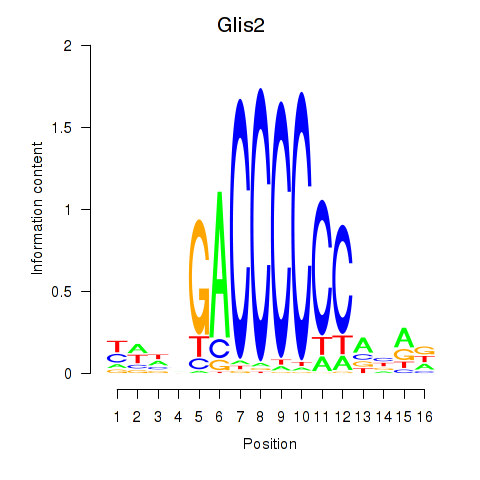
Activity profile of Glis2 motif
Sorted Z-values of Glis2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_128237087 | 7.79 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_-_126303947 | 7.32 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr11_+_87684548 | 6.64 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr7_-_126303887 | 5.92 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr8_-_72178340 | 5.59 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr4_+_114916703 | 5.17 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chrX_+_8137372 | 5.01 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_+_8137620 | 4.76 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr7_+_44221791 | 4.61 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase |
| chr11_-_11987391 | 4.11 |
ENSMUST00000093321.12
|
Grb10
|
growth factor receptor bound protein 10 |
| chr2_-_152672535 | 3.89 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr6_+_90596123 | 3.87 |
ENSMUST00000032177.10
|
Slc41a3
|
solute carrier family 41, member 3 |
| chrX_-_106446928 | 3.41 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr4_+_140737955 | 3.33 |
ENSMUST00000071977.9
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr9_+_107852733 | 3.31 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr7_-_126014027 | 3.26 |
ENSMUST00000032968.7
ENSMUST00000206325.2 |
Cd19
|
CD19 antigen |
| chr10_+_79650496 | 3.12 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr8_+_73488496 | 2.87 |
ENSMUST00000058099.9
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr2_+_90927053 | 2.73 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_+_53410697 | 2.68 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr7_-_126398343 | 2.66 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_126398165 | 2.54 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr2_-_152673032 | 2.48 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr5_-_137531471 | 2.45 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr6_-_29212295 | 2.43 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr5_-_110987604 | 2.41 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr10_+_127157784 | 2.31 |
ENSMUST00000219511.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr2_-_154411765 | 2.27 |
ENSMUST00000103145.11
|
E2f1
|
E2F transcription factor 1 |
| chr13_-_76205115 | 2.25 |
ENSMUST00000056130.8
|
Gpr150
|
G protein-coupled receptor 150 |
| chr17_+_46961250 | 2.22 |
ENSMUST00000043464.14
|
Cul7
|
cullin 7 |
| chr14_-_71003973 | 2.14 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr5_-_137531463 | 2.11 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_+_34498836 | 2.11 |
ENSMUST00000027302.14
ENSMUST00000190122.2 |
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr2_+_70948267 | 2.10 |
ENSMUST00000028403.3
|
Cybrd1
|
cytochrome b reductase 1 |
| chr7_-_24705320 | 2.02 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr5_-_137531413 | 2.02 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_+_135945705 | 1.98 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr11_+_97340962 | 1.97 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr6_-_29212239 | 1.92 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr1_-_93729562 | 1.91 |
ENSMUST00000112890.3
|
Dtymk
|
deoxythymidylate kinase |
| chr2_+_90507552 | 1.76 |
ENSMUST00000057481.7
|
Nup160
|
nucleoporin 160 |
| chr7_+_89779564 | 1.66 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_135945798 | 1.65 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr9_+_75213570 | 1.61 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr1_-_93729650 | 1.54 |
ENSMUST00000027503.14
|
Dtymk
|
deoxythymidylate kinase |
| chr5_-_110987441 | 1.51 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr9_+_50768224 | 1.49 |
ENSMUST00000174628.8
ENSMUST00000034560.14 ENSMUST00000114437.9 ENSMUST00000175645.8 ENSMUST00000176349.8 ENSMUST00000176798.8 ENSMUST00000175640.8 |
Ppp2r1b
|
protein phosphatase 2, regulatory subunit A, beta |
| chr11_+_68989763 | 1.45 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr19_+_4203603 | 1.44 |
ENSMUST00000236632.2
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr10_+_13376745 | 1.43 |
ENSMUST00000060212.13
ENSMUST00000121465.3 |
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr7_-_141614758 | 1.43 |
ENSMUST00000211000.2
ENSMUST00000209725.2 ENSMUST00000084418.4 |
Mob2
|
MOB kinase activator 2 |
| chr2_-_164585102 | 1.38 |
ENSMUST00000103096.10
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr17_-_31731222 | 1.37 |
ENSMUST00000236665.2
|
Wdr4
|
WD repeat domain 4 |
| chr5_+_139238089 | 1.33 |
ENSMUST00000130326.8
|
Get4
|
golgi to ER traffic protein 4 |
| chr1_+_135764092 | 1.28 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr7_+_89779421 | 1.28 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_+_127345909 | 1.27 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr19_+_7245591 | 1.25 |
ENSMUST00000066646.12
|
Rcor2
|
REST corepressor 2 |
| chr17_+_26882171 | 1.22 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr16_+_57173456 | 1.15 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr3_-_95214443 | 1.13 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr8_+_3681127 | 1.10 |
ENSMUST00000159911.8
ENSMUST00000004745.9 |
Stxbp2
|
syntaxin binding protein 2 |
| chr5_-_134643805 | 1.07 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr12_-_80690573 | 1.06 |
ENSMUST00000166931.2
ENSMUST00000218364.2 |
Erh
|
ERH mRNA splicing and mitosis factor |
| chr11_-_102209767 | 1.02 |
ENSMUST00000174302.8
ENSMUST00000178839.8 ENSMUST00000006754.14 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr4_-_117772163 | 0.99 |
ENSMUST00000036156.6
|
Ipo13
|
importin 13 |
| chr19_+_6451667 | 0.98 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr6_-_4747066 | 0.95 |
ENSMUST00000090686.11
ENSMUST00000133306.8 |
Sgce
|
sarcoglycan, epsilon |
| chr7_+_89779493 | 0.90 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr12_-_114117264 | 0.88 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr10_-_22607136 | 0.87 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr7_+_18817767 | 0.86 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr4_-_118477960 | 0.84 |
ENSMUST00000071972.11
|
Cfap57
|
cilia and flagella associated protein 57 |
| chr11_+_121593219 | 0.83 |
ENSMUST00000036742.14
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr5_+_136112765 | 0.82 |
ENSMUST00000042135.14
|
Rasa4
|
RAS p21 protein activator 4 |
| chr11_-_103247150 | 0.81 |
ENSMUST00000136491.3
ENSMUST00000107023.3 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr5_+_37495801 | 0.80 |
ENSMUST00000056365.9
|
Evc2
|
EvC ciliary complex subunit 2 |
| chrX_+_84617624 | 0.79 |
ENSMUST00000048250.10
ENSMUST00000137438.2 ENSMUST00000146063.2 |
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr2_+_174171860 | 0.79 |
ENSMUST00000109087.8
ENSMUST00000109084.8 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr2_+_174171799 | 0.76 |
ENSMUST00000109085.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr7_-_29892523 | 0.72 |
ENSMUST00000108196.8
|
Capns1
|
calpain, small subunit 1 |
| chr7_+_28151370 | 0.71 |
ENSMUST00000190954.7
|
Lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr6_-_116693849 | 0.71 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr3_+_127584251 | 0.70 |
ENSMUST00000164447.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr6_+_17307639 | 0.69 |
ENSMUST00000115453.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr8_-_120828211 | 0.68 |
ENSMUST00000034280.9
|
Zdhhc7
|
zinc finger, DHHC domain containing 7 |
| chr3_+_127584449 | 0.65 |
ENSMUST00000171621.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr16_+_57173632 | 0.62 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr7_-_44665639 | 0.62 |
ENSMUST00000085383.11
|
Scaf1
|
SR-related CTD-associated factor 1 |
| chr12_-_84819860 | 0.62 |
ENSMUST00000021668.10
|
Npc2
|
NPC intracellular cholesterol transporter 2 |
| chr11_+_76795346 | 0.61 |
ENSMUST00000072633.4
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr5_+_101912939 | 0.61 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr19_-_40371016 | 0.60 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_-_191776840 | 0.59 |
ENSMUST00000195815.3
|
Traf5
|
TNF receptor-associated factor 5 |
| chr7_+_99030621 | 0.59 |
ENSMUST00000037528.10
|
Gdpd5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr6_-_68609426 | 0.57 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chrX_+_149830166 | 0.57 |
ENSMUST00000026296.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr16_+_44586085 | 0.56 |
ENSMUST00000057488.15
|
Cd200r1
|
CD200 receptor 1 |
| chr19_+_11382092 | 0.56 |
ENSMUST00000153546.8
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr8_-_85413707 | 0.55 |
ENSMUST00000238301.2
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr16_-_31918485 | 0.53 |
ENSMUST00000189013.8
ENSMUST00000155966.8 ENSMUST00000096109.11 ENSMUST00000232321.2 |
Pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr4_-_49593875 | 0.51 |
ENSMUST00000151542.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr2_-_168607166 | 0.50 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr16_+_16801246 | 0.50 |
ENSMUST00000232611.2
ENSMUST00000069107.14 |
Mapk1
|
mitogen-activated protein kinase 1 |
| chr14_+_63673843 | 0.49 |
ENSMUST00000121288.2
|
Fam167a
|
family with sequence similarity 167, member A |
| chr4_-_58499398 | 0.48 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr1_-_134883645 | 0.48 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr19_+_53131187 | 0.48 |
ENSMUST00000050096.15
ENSMUST00000237832.2 |
Add3
|
adducin 3 (gamma) |
| chr1_-_118239146 | 0.48 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr19_+_8713156 | 0.45 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr7_+_101010447 | 0.45 |
ENSMUST00000137384.8
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr6_-_51446752 | 0.45 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr5_+_115644727 | 0.45 |
ENSMUST00000067268.15
ENSMUST00000086523.7 ENSMUST00000212819.3 |
Pxn
|
paxillin |
| chr3_+_105778174 | 0.44 |
ENSMUST00000164730.2
ENSMUST00000010279.10 |
Adora3
Tmigd3
|
adenosine A3 receptor transmembrane and immunoglobulin domain containing 3 |
| chr4_-_62278673 | 0.43 |
ENSMUST00000084527.10
ENSMUST00000098033.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr6_+_120643323 | 0.42 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr5_+_120727068 | 0.42 |
ENSMUST00000069259.9
ENSMUST00000094391.6 |
Iqcd
|
IQ motif containing D |
| chr1_-_96799832 | 0.42 |
ENSMUST00000071985.6
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr12_+_73170483 | 0.40 |
ENSMUST00000187549.7
ENSMUST00000021523.7 |
Mnat1
|
menage a trois 1 |
| chr5_-_124490296 | 0.40 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr5_+_136112817 | 0.38 |
ENSMUST00000100570.10
|
Rasa4
|
RAS p21 protein activator 4 |
| chr13_-_13568106 | 0.37 |
ENSMUST00000021738.10
ENSMUST00000220628.2 |
Gpr137b
|
G protein-coupled receptor 137B |
| chr1_-_152642032 | 0.37 |
ENSMUST00000111859.8
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr19_+_4198615 | 0.35 |
ENSMUST00000123874.8
ENSMUST00000008893.9 ENSMUST00000237125.2 |
Coro1b
|
coronin, actin binding protein 1B |
| chr18_+_34910064 | 0.35 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr7_-_35346553 | 0.34 |
ENSMUST00000120714.2
|
Pdcd5
|
programmed cell death 5 |
| chr3_-_90416757 | 0.32 |
ENSMUST00000107343.8
ENSMUST00000001043.14 ENSMUST00000107344.8 ENSMUST00000076639.11 ENSMUST00000107346.8 ENSMUST00000146740.8 ENSMUST00000107342.2 ENSMUST00000049937.13 |
Chtop
|
chromatin target of PRMT1 |
| chr7_+_28580847 | 0.32 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr12_-_8589545 | 0.30 |
ENSMUST00000095863.10
ENSMUST00000165657.3 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr6_+_113581708 | 0.30 |
ENSMUST00000035725.7
|
Brk1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr19_+_37538843 | 0.30 |
ENSMUST00000066439.8
ENSMUST00000238817.2 |
Exoc6
|
exocyst complex component 6 |
| chr11_+_70416185 | 0.30 |
ENSMUST00000018430.7
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr3_+_129974531 | 0.30 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr5_-_38316296 | 0.30 |
ENSMUST00000201415.4
|
Nsg1
|
neuron specific gene family member 1 |
| chr1_-_152642073 | 0.28 |
ENSMUST00000111857.3
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr6_+_42716305 | 0.28 |
ENSMUST00000213997.2
|
Olfr453
|
olfactory receptor 453 |
| chr11_-_102120917 | 0.28 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr4_-_62278761 | 0.25 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr9_-_20247390 | 0.25 |
ENSMUST00000212943.4
ENSMUST00000086473.4 |
Olfr18
|
olfactory receptor 18 |
| chr6_-_51446850 | 0.25 |
ENSMUST00000069949.13
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr19_+_29388312 | 0.24 |
ENSMUST00000112576.4
|
Pdcd1lg2
|
programmed cell death 1 ligand 2 |
| chr8_+_70953766 | 0.22 |
ENSMUST00000127983.2
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr2_+_112092271 | 0.22 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr7_-_29894471 | 0.22 |
ENSMUST00000126116.3
|
Capns1
|
calpain, small subunit 1 |
| chr10_+_126914755 | 0.22 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr7_-_92523396 | 0.21 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr7_+_5023552 | 0.21 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr2_+_174171979 | 0.21 |
ENSMUST00000109083.2
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr19_-_24878561 | 0.21 |
ENSMUST00000058600.4
|
Foxd4
|
forkhead box D4 |
| chr11_-_73781597 | 0.21 |
ENSMUST00000216608.2
ENSMUST00000214284.2 |
Olfr394
|
olfactory receptor 394 |
| chr14_-_8536869 | 0.20 |
ENSMUST00000238865.2
ENSMUST00000159275.4 |
Gm11100
|
predicted gene 11100 |
| chr10_+_80971054 | 0.20 |
ENSMUST00000125261.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr11_+_59432388 | 0.20 |
ENSMUST00000079476.10
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr1_-_74323536 | 0.19 |
ENSMUST00000190008.2
|
Aamp
|
angio-associated migratory protein |
| chr10_-_128047658 | 0.19 |
ENSMUST00000061995.10
|
Spryd4
|
SPRY domain containing 4 |
| chr19_-_34618135 | 0.19 |
ENSMUST00000087357.5
|
Ifit1bl2
|
interferon induced protein with tetratricopeptide repeats 1B like 2 |
| chr11_+_74150634 | 0.18 |
ENSMUST00000217016.2
ENSMUST00000214303.2 |
Olfr406
|
olfactory receptor 406 |
| chr11_+_69909659 | 0.17 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr5_-_123185029 | 0.17 |
ENSMUST00000045843.15
|
Morn3
|
MORN repeat containing 3 |
| chr12_-_11200306 | 0.17 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr2_-_152775122 | 0.15 |
ENSMUST00000099200.3
|
Foxs1
|
forkhead box S1 |
| chrX_+_72546680 | 0.15 |
ENSMUST00000033744.12
ENSMUST00000088429.8 ENSMUST00000114479.2 |
Atp2b3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr9_-_54554483 | 0.14 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr1_-_134883577 | 0.14 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_115417725 | 0.14 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr6_+_83985495 | 0.14 |
ENSMUST00000113821.8
|
Dysf
|
dysferlin |
| chr7_-_35346889 | 0.13 |
ENSMUST00000118501.8
|
Pdcd5
|
programmed cell death 5 |
| chr4_-_32602760 | 0.13 |
ENSMUST00000219644.2
ENSMUST00000056517.3 |
Gja10
|
gap junction protein, alpha 10 |
| chr5_-_109704339 | 0.12 |
ENSMUST00000198960.2
|
Crlf2
|
cytokine receptor-like factor 2 |
| chr5_-_123185073 | 0.10 |
ENSMUST00000031437.14
|
Morn3
|
MORN repeat containing 3 |
| chr7_-_121700958 | 0.09 |
ENSMUST00000139456.2
ENSMUST00000106471.9 ENSMUST00000123296.8 ENSMUST00000033157.10 |
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr17_-_23805187 | 0.09 |
ENSMUST00000227952.2
ENSMUST00000115516.11 |
Zfp13
|
zinc finger protein 13 |
| chr5_-_49682150 | 0.09 |
ENSMUST00000087395.11
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr17_+_34258411 | 0.08 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr10_+_36850532 | 0.08 |
ENSMUST00000019911.14
ENSMUST00000105510.2 |
Hdac2
|
histone deacetylase 2 |
| chr16_+_17577493 | 0.07 |
ENSMUST00000165790.9
|
Klhl22
|
kelch-like 22 |
| chr3_-_66888474 | 0.07 |
ENSMUST00000162439.8
ENSMUST00000162098.9 |
Shox2
|
short stature homeobox 2 |
| chrX_-_72442342 | 0.06 |
ENSMUST00000180787.3
|
Gm18336
|
predicted gene, 18336 |
| chr10_-_62723103 | 0.06 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr8_-_27903965 | 0.06 |
ENSMUST00000033882.10
|
Chrna6
|
cholinergic receptor, nicotinic, alpha polypeptide 6 |
| chr19_-_40371242 | 0.05 |
ENSMUST00000224583.2
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr8_-_25086976 | 0.05 |
ENSMUST00000033956.7
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr5_-_137246611 | 0.05 |
ENSMUST00000196391.5
|
Muc3a
|
mucin 3A, cell surface associated |
| chrX_-_135769285 | 0.04 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr4_+_155875629 | 0.04 |
ENSMUST00000105593.2
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr4_+_152160713 | 0.04 |
ENSMUST00000239025.2
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr12_-_31684588 | 0.04 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr19_+_45003304 | 0.04 |
ENSMUST00000039016.14
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr10_-_25076008 | 0.03 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr2_+_164497963 | 0.03 |
ENSMUST00000094344.6
|
Wfdc10
|
WAP four-disulfide core domain 10 |
| chr11_-_70560110 | 0.02 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chrX_-_151151680 | 0.02 |
ENSMUST00000070316.12
|
Gpr173
|
G-protein coupled receptor 173 |
| chr9_-_86762467 | 0.01 |
ENSMUST00000074501.12
ENSMUST00000239074.2 ENSMUST00000098495.10 ENSMUST00000036347.13 ENSMUST00000074468.13 |
Snap91
|
synaptosomal-associated protein 91 |
| chr9_-_121620150 | 0.01 |
ENSMUST00000215910.2
ENSMUST00000215477.2 ENSMUST00000163981.3 |
Hhatl
|
hedgehog acyltransferase-like |
| chr2_+_4980923 | 0.01 |
ENSMUST00000167607.8
ENSMUST00000115010.9 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.7 | 5.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.7 | 13.2 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 1.6 | 6.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 1.6 | 7.8 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.1 | 3.4 | GO:0006233 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.8 | 3.8 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.6 | 4.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.6 | 2.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.6 | 2.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.5 | 2.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.5 | 9.8 | GO:0015816 | glycine transport(GO:0015816) |
| 0.5 | 3.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.5 | 3.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.5 | 3.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 0.9 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.4 | 3.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 4.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.3 | 1.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 5.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 4.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.2 | 1.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 0.7 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.2 | 1.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 5.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 3.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.4 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 0.9 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 2.9 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 1.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 2.0 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 0.5 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.7 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 1.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 1.6 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 3.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 2.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.5 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 1.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.8 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.1 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 1.2 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 1.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 2.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.1 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.6 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 3.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.4 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0099645 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.4 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.5 | 3.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.5 | 12.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 7.7 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.5 | 2.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.5 | 5.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 4.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.4 | 2.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 3.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 5.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 2.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) dendritic spine neck(GO:0044326) |
| 0.2 | 3.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.4 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 1.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 7.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 4.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 2.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 1.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 7.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.7 | 4.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.7 | 2.9 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.7 | 9.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.7 | 3.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.7 | 3.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.6 | 6.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.6 | 13.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.6 | 2.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.5 | 5.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 1.4 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.4 | 3.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.4 | 3.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 1.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 1.4 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.3 | 5.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 2.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 0.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 7.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 2.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 1.0 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 2.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 2.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 1.8 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 4.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.7 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.4 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.1 | 0.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 3.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 6.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 6.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 4.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 4.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 1.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 2.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.9 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 2.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 6.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 4.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 7.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 3.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 6.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.3 | 4.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 3.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 9.9 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 4.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 9.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 5.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 3.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 3.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 3.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 2.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |