Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
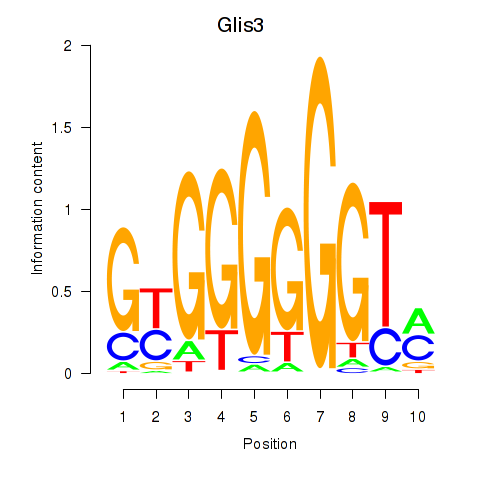
Results for Glis3
Z-value: 1.05
Transcription factors associated with Glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis3
|
ENSMUSG00000052942.14 | Glis3 |
|
Glis3
|
ENSMUSG00000052942.14 | Glis3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis3 | mm39_v1_chr19_-_28657477_28657523 | -0.55 | 4.9e-04 | Click! |
Activity profile of Glis3 motif
Sorted Z-values of Glis3 motif
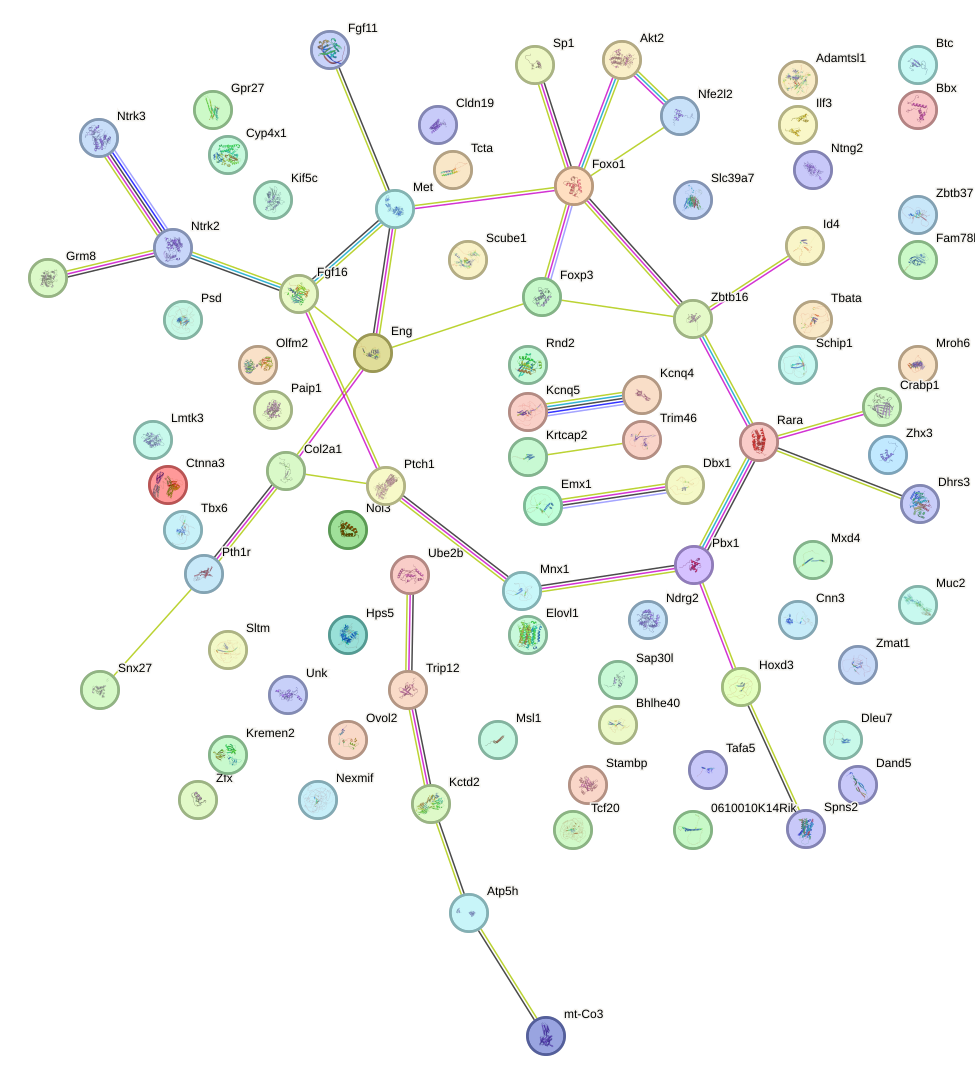
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_55798517 | 4.03 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr14_+_55798362 | 3.86 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr14_+_55797934 | 3.63 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_+_144619397 | 3.16 |
ENSMUST00000105744.8
ENSMUST00000171001.8 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_144619647 | 3.10 |
ENSMUST00000154208.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_144619696 | 2.98 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr11_+_101358990 | 2.34 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr9_-_110571645 | 2.29 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr14_-_52151026 | 1.75 |
ENSMUST00000228164.2
|
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr2_-_160714473 | 1.62 |
ENSMUST00000103111.9
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr13_+_120151982 | 1.42 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr5_-_34345014 | 1.34 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr9_-_108183356 | 1.33 |
ENSMUST00000192886.6
|
Tcta
|
T cell leukemia translocation altered gene |
| chr11_+_57692399 | 1.31 |
ENSMUST00000020826.6
|
Sap30l
|
SAP30-like |
| chr6_+_17463748 | 1.27 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr13_-_63721412 | 1.19 |
ENSMUST00000195106.2
|
Ptch1
|
patched 1 |
| chr2_-_29142965 | 1.18 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr2_-_168583670 | 1.15 |
ENSMUST00000029060.11
|
Atp9a
|
ATPase, class II, type 9A |
| chr2_-_75534985 | 1.10 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr2_-_168583817 | 1.08 |
ENSMUST00000109176.8
ENSMUST00000178504.8 |
Atp9a
|
ATPase, class II, type 9A |
| chr5_-_91550853 | 1.07 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr15_+_87428483 | 1.02 |
ENSMUST00000230414.2
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr2_-_160714904 | 1.01 |
ENSMUST00000109460.8
ENSMUST00000127201.2 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr13_+_58956495 | 1.01 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr6_+_17463925 | 0.99 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr13_+_58956077 | 0.98 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr10_-_68114543 | 0.95 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr15_-_50753792 | 0.95 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr2_-_168584020 | 0.93 |
ENSMUST00000109177.8
|
Atp9a
|
ATPase, class II, type 9A |
| chr2_-_160714749 | 0.92 |
ENSMUST00000176141.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr17_-_34250474 | 0.91 |
ENSMUST00000171872.3
ENSMUST00000025186.16 |
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr9_-_108183162 | 0.91 |
ENSMUST00000044725.9
|
Tcta
|
T cell leukemia translocation altered gene |
| chr7_+_46445512 | 0.89 |
ENSMUST00000006774.11
ENSMUST00000165031.8 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr17_-_34250616 | 0.87 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr13_+_120151915 | 0.85 |
ENSMUST00000225543.2
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr1_-_168259710 | 0.84 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr11_-_72380730 | 0.84 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr1_-_168259264 | 0.81 |
ENSMUST00000176790.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr15_-_82783978 | 0.79 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20 |
| chr6_+_17463819 | 0.79 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr2_-_73722932 | 0.78 |
ENSMUST00000154456.8
ENSMUST00000090802.11 ENSMUST00000055833.12 ENSMUST00000112007.8 ENSMUST00000112016.9 |
Atf2
|
activating transcription factor 2 |
| chr9_-_108183140 | 0.78 |
ENSMUST00000195615.2
|
Tcta
|
T cell leukemia translocation altered gene |
| chr16_+_20492014 | 0.77 |
ENSMUST00000154950.8
ENSMUST00000115461.8 ENSMUST00000136713.5 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr7_+_45434755 | 0.77 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr7_-_46445085 | 0.70 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr3_+_52175757 | 0.70 |
ENSMUST00000053764.7
|
Foxo1
|
forkhead box O1 |
| chr2_+_32536594 | 0.69 |
ENSMUST00000113272.8
ENSMUST00000009705.14 ENSMUST00000167841.8 |
Eng
|
endoglin |
| chr3_+_121220146 | 0.69 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr7_-_46445305 | 0.67 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr9_+_21279802 | 0.65 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr7_-_49286594 | 0.65 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr11_+_98818640 | 0.64 |
ENSMUST00000107474.8
|
Rara
|
retinoic acid receptor, alpha |
| chr13_+_58955675 | 0.64 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr6_+_85164420 | 0.64 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chr2_-_144173615 | 0.63 |
ENSMUST00000103171.10
|
Ovol2
|
ovo like zinc finger 2 |
| chr12_+_102915709 | 0.63 |
ENSMUST00000179002.8
|
Unc79
|
unc-79 homolog |
| chr15_+_102314578 | 0.63 |
ENSMUST00000170884.8
ENSMUST00000163709.8 |
Sp1
|
trans-acting transcription factor 1 |
| chr2_-_73722874 | 0.62 |
ENSMUST00000136958.8
ENSMUST00000112010.9 ENSMUST00000128531.8 ENSMUST00000112017.8 |
Atf2
|
activating transcription factor 2 |
| chrM_+_8603 | 0.60 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr11_-_115310743 | 0.59 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr17_-_23964807 | 0.59 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr1_-_160862364 | 0.58 |
ENSMUST00000177003.2
ENSMUST00000159250.9 ENSMUST00000162226.9 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr4_-_114991174 | 0.54 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr1_-_168259465 | 0.54 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr8_+_109441276 | 0.53 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr9_-_20638233 | 0.52 |
ENSMUST00000217198.2
|
Olfm2
|
olfactomedin 2 |
| chrX_-_37653396 | 0.51 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chrX_+_104807868 | 0.51 |
ENSMUST00000033581.4
|
Fgf16
|
fibroblast growth factor 16 |
| chr1_-_168259070 | 0.50 |
ENSMUST00000064438.11
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr7_+_126380655 | 0.50 |
ENSMUST00000172352.8
ENSMUST00000094037.5 |
Tbx6
|
T-box 6 |
| chr7_-_78228116 | 0.49 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr4_+_118285275 | 0.49 |
ENSMUST00000006557.13
ENSMUST00000167636.8 ENSMUST00000102673.11 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr15_+_102315579 | 0.46 |
ENSMUST00000169619.2
|
Sp1
|
trans-acting transcription factor 1 |
| chr16_-_50252703 | 0.46 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr10_+_63293284 | 0.46 |
ENSMUST00000105440.8
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr13_+_119565424 | 0.46 |
ENSMUST00000026520.14
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr11_+_115310963 | 0.46 |
ENSMUST00000106533.8
ENSMUST00000123345.2 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr1_+_166828982 | 0.46 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr8_+_106002772 | 0.46 |
ENSMUST00000014920.8
|
Nol3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr2_+_49509288 | 0.45 |
ENSMUST00000028102.14
|
Kif5c
|
kinesin family member 5C |
| chr5_-_8417982 | 0.45 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr4_+_85972125 | 0.44 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr14_-_62530408 | 0.43 |
ENSMUST00000063169.10
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr7_+_27290969 | 0.43 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr1_+_166829001 | 0.42 |
ENSMUST00000126198.3
|
Fam78b
|
family with sequence similarity 78, member B |
| chr6_-_28134544 | 0.42 |
ENSMUST00000115323.8
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr11_+_115310885 | 0.41 |
ENSMUST00000103035.10
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr11_-_69692542 | 0.40 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chr11_-_70128587 | 0.40 |
ENSMUST00000108576.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr7_+_27291126 | 0.39 |
ENSMUST00000167435.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chrX_-_93166693 | 0.39 |
ENSMUST00000113925.8
|
Zfx
|
zinc finger protein X-linked |
| chr11_-_88608958 | 0.39 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr3_+_68401536 | 0.38 |
ENSMUST00000182719.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr17_-_81035453 | 0.36 |
ENSMUST00000234133.2
ENSMUST00000112389.9 ENSMUST00000025089.9 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr15_-_83609127 | 0.36 |
ENSMUST00000171496.9
ENSMUST00000043634.12 ENSMUST00000076060.12 ENSMUST00000016907.8 |
Scube1
|
signal peptide, CUB domain, EGF-like 1 |
| chr9_+_54672032 | 0.36 |
ENSMUST00000034830.9
|
Crabp1
|
cellular retinoic acid binding protein I |
| chr1_-_22031718 | 0.35 |
ENSMUST00000029667.13
ENSMUST00000173058.8 ENSMUST00000173404.2 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr4_-_120604445 | 0.35 |
ENSMUST00000030376.8
|
Kcnq4
|
potassium voltage-gated channel, subfamily Q, member 4 |
| chr9_-_48747474 | 0.35 |
ENSMUST00000216150.2
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr6_+_99669640 | 0.34 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr2_+_74566740 | 0.34 |
ENSMUST00000111982.8
|
Hoxd3
|
homeobox D3 |
| chr1_-_84816379 | 0.34 |
ENSMUST00000187818.2
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr6_+_108637577 | 0.33 |
ENSMUST00000032194.11
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr7_-_109271171 | 0.33 |
ENSMUST00000208734.2
|
Denn2b
|
DENN domain containing 2B |
| chrX_+_7439839 | 0.33 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr7_+_141276575 | 0.32 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr6_-_83549399 | 0.32 |
ENSMUST00000206592.2
ENSMUST00000206400.2 |
Stambp
|
STAM binding protein |
| chr15_-_97902576 | 0.32 |
ENSMUST00000023123.15
|
Col2a1
|
collagen, type II, alpha 1 |
| chr19_-_46314945 | 0.31 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr9_-_48747232 | 0.30 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr13_+_48414582 | 0.29 |
ENSMUST00000021810.3
|
Id4
|
inhibitor of DNA binding 4 |
| chr11_-_51891259 | 0.29 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr13_+_119565669 | 0.29 |
ENSMUST00000173627.8
ENSMUST00000126957.9 ENSMUST00000174691.8 |
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr5_-_29683468 | 0.29 |
ENSMUST00000165512.4
ENSMUST00000001608.8 |
Mnx1
|
motor neuron and pancreas homeobox 1 |
| chrX_-_103244728 | 0.27 |
ENSMUST00000056502.7
|
Nexmif
|
neurite extension and migration factor |
| chr13_+_83720484 | 0.27 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_115921129 | 0.27 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr15_-_97902515 | 0.26 |
ENSMUST00000088355.12
|
Col2a1
|
collagen, type II, alpha 1 |
| chr3_+_89153258 | 0.26 |
ENSMUST00000040888.12
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr10_+_61010983 | 0.25 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr18_-_78249612 | 0.25 |
ENSMUST00000163367.3
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr4_+_119112590 | 0.25 |
ENSMUST00000084309.12
|
Cldn19
|
claudin 19 |
| chr4_-_114991478 | 0.25 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr15_-_75760602 | 0.25 |
ENSMUST00000184858.2
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr3_-_94490023 | 0.24 |
ENSMUST00000029783.16
|
Snx27
|
sorting nexin family member 27 |
| chr4_+_119112692 | 0.24 |
ENSMUST00000094823.4
|
Cldn19
|
claudin 19 |
| chr13_+_119565118 | 0.23 |
ENSMUST00000109203.9
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chrX_-_133909934 | 0.23 |
ENSMUST00000113185.9
ENSMUST00000064659.6 |
Zmat1
|
zinc finger, matrin type 1 |
| chr15_-_50753437 | 0.23 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr3_-_89153135 | 0.22 |
ENSMUST00000041022.15
|
Trim46
|
tripartite motif-containing 46 |
| chr11_+_98686617 | 0.22 |
ENSMUST00000107485.8
ENSMUST00000107487.10 |
Msl1
|
male specific lethal 1 |
| chr10_-_62723103 | 0.22 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chrX_-_110606766 | 0.21 |
ENSMUST00000113422.9
ENSMUST00000038472.7 |
Hdx
|
highly divergent homeobox |
| chr7_+_5023375 | 0.21 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
| chr8_-_88686188 | 0.21 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chrX_-_93166964 | 0.21 |
ENSMUST00000137853.8
|
Zfx
|
zinc finger protein X-linked |
| chr17_-_25652750 | 0.20 |
ENSMUST00000159610.8
ENSMUST00000159048.8 ENSMUST00000078496.12 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chrX_-_154406321 | 0.20 |
ENSMUST00000038665.6
|
Ptchd1
|
patched domain containing 1 |
| chr2_-_152793376 | 0.20 |
ENSMUST00000123121.9
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr13_+_38009951 | 0.20 |
ENSMUST00000138043.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr13_-_30168374 | 0.19 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr11_-_106240215 | 0.19 |
ENSMUST00000021056.8
|
Scn4a
|
sodium channel, voltage-gated, type IV, alpha |
| chr18_-_47501513 | 0.19 |
ENSMUST00000076043.13
ENSMUST00000135790.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr13_+_83720457 | 0.18 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_144174066 | 0.18 |
ENSMUST00000037423.4
|
Ovol2
|
ovo like zinc finger 2 |
| chr15_+_89383799 | 0.17 |
ENSMUST00000109309.9
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr19_-_47303184 | 0.17 |
ENSMUST00000237796.2
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr5_-_134485081 | 0.16 |
ENSMUST00000111244.5
|
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr2_-_152793469 | 0.16 |
ENSMUST00000037715.7
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr7_+_18659787 | 0.16 |
ENSMUST00000032571.10
ENSMUST00000220302.2 |
Nova2
|
NOVA alternative splicing regulator 2 |
| chr15_+_89384317 | 0.15 |
ENSMUST00000135214.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr2_-_122199604 | 0.15 |
ENSMUST00000151130.8
|
Shf
|
Src homology 2 domain containing F |
| chr15_-_103248512 | 0.15 |
ENSMUST00000168828.3
|
Zfp385a
|
zinc finger protein 385A |
| chr3_-_142587678 | 0.15 |
ENSMUST00000043812.15
|
Pkn2
|
protein kinase N2 |
| chr17_-_24866749 | 0.14 |
ENSMUST00000234121.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr7_-_142453722 | 0.14 |
ENSMUST00000000219.10
|
Th
|
tyrosine hydroxylase |
| chr6_-_52222776 | 0.13 |
ENSMUST00000048026.10
|
Hoxa11
|
homeobox A11 |
| chr9_-_50663648 | 0.12 |
ENSMUST00000217159.2
|
Hspb2
|
heat shock protein 2 |
| chr11_-_70128462 | 0.12 |
ENSMUST00000100950.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr11_+_105072353 | 0.12 |
ENSMUST00000106941.9
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr1_-_168259839 | 0.11 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr14_+_53257873 | 0.11 |
ENSMUST00000196756.2
|
Trav7d-6
|
T cell receptor alpha variable 7D-6 |
| chr7_+_3381434 | 0.11 |
ENSMUST00000092891.6
|
Cacng7
|
calcium channel, voltage-dependent, gamma subunit 7 |
| chr2_-_152793607 | 0.11 |
ENSMUST00000109811.4
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr15_+_102314809 | 0.10 |
ENSMUST00000001326.7
|
Sp1
|
trans-acting transcription factor 1 |
| chrX_+_98864627 | 0.10 |
ENSMUST00000096363.3
|
Tmem28
|
transmembrane protein 28 |
| chr11_-_33097400 | 0.10 |
ENSMUST00000020507.8
|
Fgf18
|
fibroblast growth factor 18 |
| chr17_+_35191661 | 0.10 |
ENSMUST00000007248.5
|
Hspa1l
|
heat shock protein 1-like |
| chr4_-_148215135 | 0.09 |
ENSMUST00000030862.5
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr1_-_74323795 | 0.09 |
ENSMUST00000178235.8
ENSMUST00000006462.14 |
Aamp
|
angio-associated migratory protein |
| chr2_-_122199643 | 0.08 |
ENSMUST00000125826.8
|
Shf
|
Src homology 2 domain containing F |
| chr11_+_96162283 | 0.08 |
ENSMUST00000000010.9
ENSMUST00000174042.3 |
Hoxb9
|
homeobox B9 |
| chr9_+_107805647 | 0.08 |
ENSMUST00000085073.2
|
Actl11
|
actin-like 11 |
| chr18_-_47501897 | 0.07 |
ENSMUST00000019791.14
ENSMUST00000115449.9 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr9_+_106247943 | 0.06 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr19_+_41471395 | 0.06 |
ENSMUST00000237208.2
ENSMUST00000238398.2 |
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr17_-_85097945 | 0.05 |
ENSMUST00000112308.9
|
Lrpprc
|
leucine-rich PPR-motif containing |
| chr9_+_31191820 | 0.05 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chr16_+_22713593 | 0.05 |
ENSMUST00000232674.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr10_-_43934774 | 0.04 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr1_-_74323536 | 0.04 |
ENSMUST00000190008.2
|
Aamp
|
angio-associated migratory protein |
| chr6_+_108637816 | 0.04 |
ENSMUST00000163617.2
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr13_-_40883893 | 0.04 |
ENSMUST00000021787.7
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr9_+_50663171 | 0.03 |
ENSMUST00000214609.2
|
Cryab
|
crystallin, alpha B |
| chr9_-_50663571 | 0.03 |
ENSMUST00000042790.5
|
Hspb2
|
heat shock protein 2 |
| chr8_+_70953766 | 0.03 |
ENSMUST00000127983.2
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr15_-_94302139 | 0.01 |
ENSMUST00000035342.11
ENSMUST00000155907.2 |
Adamts20
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 20 |
| chr2_-_119308094 | 0.01 |
ENSMUST00000110808.2
ENSMUST00000049920.14 |
Ino80
|
INO80 complex subunit |
| chr16_+_20492267 | 0.01 |
ENSMUST00000115460.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.4 | 3.0 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 9.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 1.2 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.4 | 1.1 | GO:1904753 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.4 | 1.4 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.3 | 1.8 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.2 | 3.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 1.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.2 | 0.7 | GO:0003032 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.2 | 0.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.5 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.8 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 2.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 2.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 0.5 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.4 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.5 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 0.3 | GO:0002660 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.8 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.4 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.4 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 2.8 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.6 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 1.1 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 0.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 1.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.3 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 3.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.6 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.6 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.7 | GO:0071455 | response to hyperoxia(GO:0055093) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.6 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.4 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 1.2 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.6 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 1.9 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0033128 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.0 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 1.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.4 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.5 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 1.4 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.3 | 12.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 3.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 4.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 3.0 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.8 | 2.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.4 | 2.6 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.3 | 2.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 1.2 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 3.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.7 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.5 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.1 | 1.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.4 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 1.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.3 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 3.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.7 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |