Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Gsc2_Dmbx1
Z-value: 0.44
Transcription factors associated with Gsc2_Dmbx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
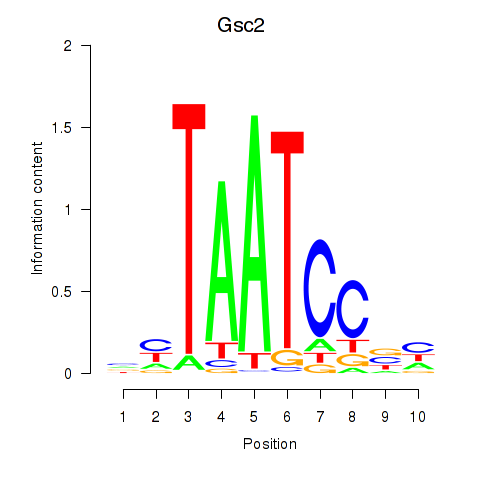
Gsc2
|
ENSMUSG00000022738.8 | Gsc2 |
|
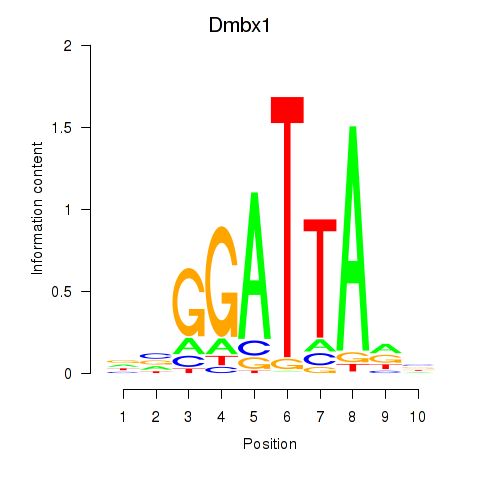
Dmbx1
|
ENSMUSG00000028707.16 | Dmbx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gsc2 | mm39_v1_chr16_-_17732923_17732923 | 0.16 | 3.4e-01 | Click! |
| Dmbx1 | mm39_v1_chr4_-_115797118_115797129 | -0.10 | 5.8e-01 | Click! |
Activity profile of Gsc2_Dmbx1 motif
Sorted Z-values of Gsc2_Dmbx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsc2_Dmbx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_68864666 | 0.75 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr14_+_26722319 | 0.59 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr14_+_36776775 | 0.49 |
ENSMUST00000120052.2
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chrX_+_73468140 | 0.47 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr3_+_28835425 | 0.46 |
ENSMUST00000060500.9
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr14_-_118289557 | 0.46 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr11_+_94901104 | 0.37 |
ENSMUST00000124735.2
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr11_-_70405429 | 0.37 |
ENSMUST00000021179.4
|
Vmo1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr6_+_119213541 | 0.33 |
ENSMUST00000186622.3
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr6_-_29380423 | 0.33 |
ENSMUST00000147483.3
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr6_+_119213468 | 0.33 |
ENSMUST00000238905.2
ENSMUST00000037434.13 |
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr6_-_29380467 | 0.30 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr6_-_124817155 | 0.30 |
ENSMUST00000024206.6
|
Gnb3
|
guanine nucleotide binding protein (G protein), beta 3 |
| chr6_+_3993774 | 0.29 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr11_+_97917520 | 0.28 |
ENSMUST00000092425.11
|
Rpl19
|
ribosomal protein L19 |
| chr1_+_82817794 | 0.27 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chrM_+_8603 | 0.25 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr19_+_4189786 | 0.25 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr7_+_43361930 | 0.25 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr6_+_136931519 | 0.24 |
ENSMUST00000137768.2
|
Pde6h
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr1_-_172958803 | 0.24 |
ENSMUST00000073663.3
|
Olfr1408
|
olfactory receptor 1408 |
| chr4_-_139974062 | 0.24 |
ENSMUST00000039331.9
|
Igsf21
|
immunoglobulin superfamily, member 21 |
| chr11_+_78213791 | 0.23 |
ENSMUST00000017534.15
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr2_+_121287444 | 0.23 |
ENSMUST00000126764.2
|
Hypk
|
huntingtin interacting protein K |
| chr1_-_132294807 | 0.22 |
ENSMUST00000136828.3
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr9_-_108443916 | 0.21 |
ENSMUST00000194381.2
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr17_+_34124078 | 0.21 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr15_+_4404965 | 0.21 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr13_-_64645606 | 0.21 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr13_+_23936250 | 0.21 |
ENSMUST00000091703.3
|
H3c2
|
H3 clustered histone 2 |
| chr17_-_28584117 | 0.20 |
ENSMUST00000233420.2
|
Tulp1
|
tubby like protein 1 |
| chr2_+_86128161 | 0.20 |
ENSMUST00000054746.5
|
Olfr1052
|
olfactory receptor 1052 |
| chr6_+_115111860 | 0.20 |
ENSMUST00000169345.4
|
Syn2
|
synapsin II |
| chrX_-_166907286 | 0.20 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr13_-_21967540 | 0.20 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11 |
| chr9_+_50528608 | 0.20 |
ENSMUST00000000171.15
ENSMUST00000151197.8 |
Pih1d2
|
PIH1 domain containing 2 |
| chr3_-_144555062 | 0.19 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr4_+_80752535 | 0.19 |
ENSMUST00000102831.8
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr2_-_23939401 | 0.19 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr4_+_136349924 | 0.19 |
ENSMUST00000178843.3
|
Lactbl1
|
lactamase, beta-like 1 |
| chr14_+_99536111 | 0.19 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5 |
| chr3_+_66127330 | 0.18 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr12_-_101943134 | 0.18 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr6_+_115111872 | 0.18 |
ENSMUST00000009538.12
ENSMUST00000203450.2 |
Syn2
|
synapsin II |
| chrX_+_7594670 | 0.18 |
ENSMUST00000033489.8
|
Praf2
|
PRA1 domain family 2 |
| chr10_-_22025169 | 0.17 |
ENSMUST00000179054.8
ENSMUST00000069372.7 |
E030030I06Rik
|
RIKEN cDNA E030030I06 gene |
| chr2_-_9888804 | 0.17 |
ENSMUST00000114915.3
|
9230102O04Rik
|
RIKEN cDNA 9230102O04 gene |
| chr7_-_4517608 | 0.17 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr10_+_115854118 | 0.17 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_+_50528813 | 0.16 |
ENSMUST00000141366.8
|
Pih1d2
|
PIH1 domain containing 2 |
| chr6_+_115911302 | 0.16 |
ENSMUST00000203877.2
|
Rho
|
rhodopsin |
| chr4_+_80752360 | 0.16 |
ENSMUST00000133655.8
ENSMUST00000006151.13 |
Tyrp1
|
tyrosinase-related protein 1 |
| chr3_-_64197130 | 0.15 |
ENSMUST00000233531.2
ENSMUST00000176328.9 |
Vmn2r3
|
vomeronasal 2, receptor 3 |
| chr7_-_4755971 | 0.15 |
ENSMUST00000183971.8
ENSMUST00000182173.2 ENSMUST00000182738.8 ENSMUST00000182111.8 ENSMUST00000184143.8 ENSMUST00000182048.2 ENSMUST00000063324.14 |
Cox6b2
|
cytochrome c oxidase subunit 6B2 |
| chr19_+_36061096 | 0.14 |
ENSMUST00000025714.9
|
Rpp30
|
ribonuclease P/MRP 30 subunit |
| chr4_-_140780972 | 0.14 |
ENSMUST00000040222.14
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr17_-_23892798 | 0.14 |
ENSMUST00000047436.11
ENSMUST00000115490.9 ENSMUST00000095579.11 |
Thoc6
|
THO complex 6 |
| chr1_-_154692678 | 0.14 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr7_-_4517367 | 0.14 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr8_+_13455080 | 0.14 |
ENSMUST00000033827.8
ENSMUST00000209909.2 |
Grk1
|
G protein-coupled receptor kinase 1 |
| chr17_+_47221377 | 0.13 |
ENSMUST00000024773.6
|
Prph2
|
peripherin 2 |
| chr7_-_4517559 | 0.13 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chrX_+_133486391 | 0.13 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr15_+_18819033 | 0.13 |
ENSMUST00000166873.9
|
Cdh10
|
cadherin 10 |
| chr9_+_32305259 | 0.13 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr7_+_13132040 | 0.13 |
ENSMUST00000005791.14
|
Cabp5
|
calcium binding protein 5 |
| chr1_-_44258112 | 0.13 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chrX_-_42256694 | 0.12 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr7_+_57069417 | 0.12 |
ENSMUST00000085240.11
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr11_-_69127848 | 0.12 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr13_-_116446166 | 0.11 |
ENSMUST00000036060.13
|
Isl1
|
ISL1 transcription factor, LIM/homeodomain |
| chr17_-_74602469 | 0.11 |
ENSMUST00000233144.2
|
Memo1
|
mediator of cell motility 1 |
| chr17_+_34092811 | 0.11 |
ENSMUST00000226885.2
|
Smim40-ps
|
small integral membrane protein 40, pseudogene |
| chr1_-_160405464 | 0.11 |
ENSMUST00000238289.2
|
Gpr52
|
G protein-coupled receptor 52 |
| chr8_-_114575247 | 0.11 |
ENSMUST00000093113.5
|
Adamts18
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 18 |
| chr14_-_55762416 | 0.10 |
ENSMUST00000178694.3
|
Nrl
|
neural retina leucine zipper gene |
| chr4_+_19280850 | 0.10 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr18_-_78222349 | 0.10 |
ENSMUST00000237290.2
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr14_-_55762395 | 0.10 |
ENSMUST00000228287.2
|
Nrl
|
neural retina leucine zipper gene |
| chr14_-_55762432 | 0.10 |
ENSMUST00000062232.15
|
Nrl
|
neural retina leucine zipper gene |
| chr7_-_127324788 | 0.10 |
ENSMUST00000076091.4
|
Ctf2
|
cardiotrophin 2 |
| chrX_+_159551009 | 0.10 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr14_-_20027112 | 0.10 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr3_-_144804784 | 0.10 |
ENSMUST00000040465.11
ENSMUST00000198993.2 |
Clca2
|
chloride channel accessory 2 |
| chrX_+_133195974 | 0.10 |
ENSMUST00000037687.8
|
Tmem35a
|
transmembrane protein 35A |
| chr16_-_17732923 | 0.10 |
ENSMUST00000012279.6
ENSMUST00000232493.2 |
Gsc2
|
goosecoid homebox 2 |
| chr10_-_18890281 | 0.10 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr15_-_74606162 | 0.10 |
ENSMUST00000023260.5
|
Lypd2
|
Ly6/Plaur domain containing 2 |
| chr19_-_12742811 | 0.10 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr16_+_65612083 | 0.09 |
ENSMUST00000168064.3
|
Vgll3
|
vestigial like family member 3 |
| chr17_+_27874483 | 0.09 |
ENSMUST00000114873.8
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr8_-_62576140 | 0.09 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr17_+_27874546 | 0.09 |
ENSMUST00000232437.2
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr2_+_67276338 | 0.09 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr1_-_110905082 | 0.09 |
ENSMUST00000094626.5
|
Cdh19
|
cadherin 19, type 2 |
| chrX_+_37689503 | 0.09 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr15_+_11000802 | 0.09 |
ENSMUST00000117100.4
|
Slc45a2
|
solute carrier family 45, member 2 |
| chr11_+_23206565 | 0.09 |
ENSMUST00000136235.2
|
Xpo1
|
exportin 1 |
| chr19_-_47680528 | 0.08 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr17_+_27874519 | 0.08 |
ENSMUST00000045896.11
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr11_+_69886652 | 0.08 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr10_+_128061699 | 0.08 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr2_-_120916316 | 0.08 |
ENSMUST00000028721.8
|
Tgm5
|
transglutaminase 5 |
| chr4_-_139560831 | 0.08 |
ENSMUST00000030508.14
|
Pax7
|
paired box 7 |
| chr7_+_126575510 | 0.07 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr7_+_63803663 | 0.07 |
ENSMUST00000206314.2
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr11_+_70350963 | 0.07 |
ENSMUST00000126105.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr12_-_115206715 | 0.07 |
ENSMUST00000103527.2
ENSMUST00000194071.2 |
Ighv1-56
|
immunoglobulin heavy variable 1-56 |
| chr15_-_101801351 | 0.07 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr17_-_72059865 | 0.07 |
ENSMUST00000057405.9
|
Pcare
|
photoreceptor cilium actin regulator |
| chr6_+_90246088 | 0.07 |
ENSMUST00000058039.3
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr19_-_46314945 | 0.07 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr5_-_137643799 | 0.07 |
ENSMUST00000196511.2
|
Irs3
|
insulin receptor substrate 3 |
| chr6_-_38086484 | 0.07 |
ENSMUST00000114908.5
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr10_+_116822219 | 0.06 |
ENSMUST00000020378.5
|
Best3
|
bestrophin 3 |
| chr1_-_13730732 | 0.06 |
ENSMUST00000027071.7
|
Lactb2
|
lactamase, beta 2 |
| chr11_+_70350725 | 0.06 |
ENSMUST00000147289.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chrX_+_159551171 | 0.06 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr11_-_90578397 | 0.06 |
ENSMUST00000107869.9
ENSMUST00000154599.2 ENSMUST00000107868.8 ENSMUST00000020849.9 |
Tom1l1
|
target of myb1-like 1 (chicken) |
| chr7_-_132725575 | 0.06 |
ENSMUST00000171968.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr3_-_84167119 | 0.06 |
ENSMUST00000107691.8
|
Trim2
|
tripartite motif-containing 2 |
| chr11_+_67586140 | 0.06 |
ENSMUST00000021290.2
|
Rcvrn
|
recoverin |
| chr7_+_63803583 | 0.06 |
ENSMUST00000085222.12
ENSMUST00000206277.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr11_-_59078565 | 0.06 |
ENSMUST00000170895.3
|
Guk1
|
guanylate kinase 1 |
| chr11_+_19874354 | 0.06 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr11_+_69886603 | 0.06 |
ENSMUST00000018716.10
|
Phf23
|
PHD finger protein 23 |
| chr2_-_51863203 | 0.06 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr11_-_70510010 | 0.06 |
ENSMUST00000102556.10
ENSMUST00000014753.9 |
Chrne
|
cholinergic receptor, nicotinic, epsilon polypeptide |
| chr5_-_66161621 | 0.06 |
ENSMUST00000201351.4
|
9130230L23Rik
|
RIKEN cDNA 9130230L23 gene |
| chrX_+_56134672 | 0.05 |
ENSMUST00000114745.9
ENSMUST00000114746.2 |
Vgll1
|
vestigial like family member 1 |
| chr9_+_30941924 | 0.05 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr6_-_93890237 | 0.05 |
ENSMUST00000204167.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr13_+_75237939 | 0.05 |
ENSMUST00000022075.6
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr4_+_84802592 | 0.05 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr14_-_28691423 | 0.05 |
ENSMUST00000225985.2
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr10_+_23952398 | 0.05 |
ENSMUST00000051133.6
|
Taar8a
|
trace amine-associated receptor 8A |
| chr6_+_38086190 | 0.05 |
ENSMUST00000031851.5
|
Tmem213
|
transmembrane protein 213 |
| chrX_+_20291927 | 0.05 |
ENSMUST00000115384.9
|
Jade3
|
jade family PHD finger 3 |
| chr5_+_20112500 | 0.05 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_+_23206001 | 0.05 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr7_+_103684652 | 0.05 |
ENSMUST00000213214.2
|
Olfr641
|
olfactory receptor 641 |
| chr4_-_140781015 | 0.05 |
ENSMUST00000102491.10
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr19_+_12861219 | 0.05 |
ENSMUST00000049624.3
|
Olfr1445
|
olfactory receptor 1445 |
| chr2_-_93292257 | 0.05 |
ENSMUST00000150508.8
|
Cd82
|
CD82 antigen |
| chr7_-_25098122 | 0.05 |
ENSMUST00000105177.3
ENSMUST00000149349.2 |
Lipe
|
lipase, hormone sensitive |
| chr5_+_3394187 | 0.05 |
ENSMUST00000042410.5
|
Cdk6
|
cyclin-dependent kinase 6 |
| chrX_+_99615836 | 0.05 |
ENSMUST00000096361.5
|
Awat1
|
acyl-CoA wax alcohol acyltransferase 1 |
| chr2_-_29983618 | 0.04 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr11_-_5049223 | 0.04 |
ENSMUST00000079949.13
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr10_+_90412827 | 0.04 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr5_-_137643954 | 0.04 |
ENSMUST00000057314.4
|
Irs3
|
insulin receptor substrate 3 |
| chr1_-_4430481 | 0.04 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr1_-_136158027 | 0.04 |
ENSMUST00000150163.8
ENSMUST00000144464.7 |
Inava
|
innate immunity activator |
| chr7_-_19013936 | 0.04 |
ENSMUST00000032560.6
|
Ppm1n
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr7_-_126574959 | 0.04 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr17_-_59298338 | 0.03 |
ENSMUST00000025064.14
|
Pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr13_-_105430889 | 0.03 |
ENSMUST00000226044.2
|
Rnf180
|
ring finger protein 180 |
| chr14_-_20027219 | 0.03 |
ENSMUST00000055100.14
ENSMUST00000162425.8 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr12_-_115706126 | 0.03 |
ENSMUST00000166645.2
ENSMUST00000196690.2 |
Ighv1-71
|
immunoglobulin heavy variable 1-71 |
| chr13_-_105430932 | 0.03 |
ENSMUST00000224662.2
|
Rnf180
|
ring finger protein 180 |
| chr17_-_71309815 | 0.03 |
ENSMUST00000123686.8
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr19_+_8595369 | 0.03 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr3_-_9029097 | 0.02 |
ENSMUST00000091354.12
ENSMUST00000094381.11 |
Tpd52
|
tumor protein D52 |
| chr4_+_101407608 | 0.02 |
ENSMUST00000094953.11
ENSMUST00000106933.2 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr14_-_55163452 | 0.02 |
ENSMUST00000227037.2
|
Efs
|
embryonal Fyn-associated substrate |
| chr11_+_5049404 | 0.02 |
ENSMUST00000134267.8
ENSMUST00000036320.12 ENSMUST00000150632.8 |
Rhbdd3
|
rhomboid domain containing 3 |
| chr7_-_109092834 | 0.02 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chr2_+_101455079 | 0.02 |
ENSMUST00000111227.2
|
Rag2
|
recombination activating gene 2 |
| chr16_+_17051423 | 0.02 |
ENSMUST00000090190.14
ENSMUST00000232082.2 ENSMUST00000232426.2 |
Hic2
Gm49573
|
hypermethylated in cancer 2 predicted gene, 49573 |
| chr11_-_5049082 | 0.01 |
ENSMUST00000063232.7
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr17_+_28259749 | 0.01 |
ENSMUST00000233869.2
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr14_+_53100756 | 0.01 |
ENSMUST00000103616.5
ENSMUST00000186370.2 |
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr11_-_43792013 | 0.01 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr16_-_19703014 | 0.01 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr1_+_177272297 | 0.01 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr2_-_73316809 | 0.01 |
ENSMUST00000141264.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_12762712 | 0.01 |
ENSMUST00000186051.7
|
Sulf1
|
sulfatase 1 |
| chr5_+_77413282 | 0.01 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chr2_-_51862941 | 0.01 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr11_-_5049036 | 0.01 |
ENSMUST00000102930.10
ENSMUST00000093365.12 ENSMUST00000073308.11 |
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr1_-_155108455 | 0.01 |
ENSMUST00000035914.5
|
BC034090
|
cDNA sequence BC034090 |
| chrM_+_7779 | 0.00 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr19_-_4189603 | 0.00 |
ENSMUST00000025761.8
|
Cabp4
|
calcium binding protein 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.3 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.2 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0034148 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |