Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Gzf1
Z-value: 0.64
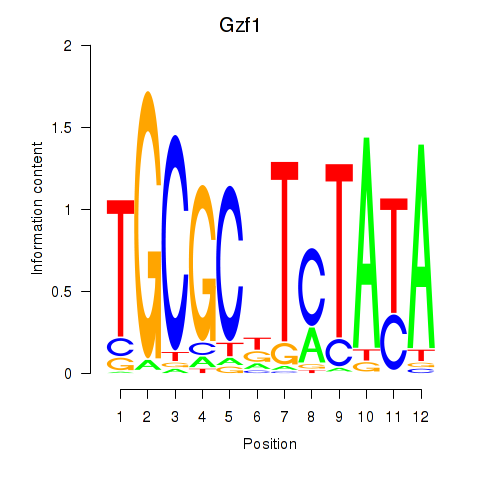
Transcription factors associated with Gzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gzf1
|
ENSMUSG00000027439.10 | Gzf1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gzf1 | mm39_v1_chr2_+_148522943_148523070 | 0.27 | 1.1e-01 | Click! |
Activity profile of Gzf1 motif
Sorted Z-values of Gzf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Gzf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_133856025 | 3.48 |
ENSMUST00000105879.2
ENSMUST00000030651.9 |
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3 |
| chr17_+_47905553 | 3.00 |
ENSMUST00000182846.3
|
Ccnd3
|
cyclin D3 |
| chr11_-_33113071 | 1.90 |
ENSMUST00000093201.13
ENSMUST00000101375.5 ENSMUST00000109354.10 ENSMUST00000075641.10 |
Npm1
|
nucleophosmin 1 |
| chr8_-_79235505 | 1.57 |
ENSMUST00000211719.2
ENSMUST00000049245.10 |
Rbmxl1
|
RNA binding motif protein, X-linked like-1 |
| chr14_+_19801333 | 1.30 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr4_+_129407374 | 1.21 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr8_+_23901506 | 1.18 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr4_-_129436465 | 1.02 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr2_+_162773440 | 0.89 |
ENSMUST00000130411.7
ENSMUST00000126163.3 |
Srsf6
|
serine and arginine-rich splicing factor 6 |
| chr12_+_85645801 | 0.74 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr7_-_30312246 | 0.63 |
ENSMUST00000006476.6
|
Upk1a
|
uroplakin 1A |
| chr1_+_180553569 | 0.60 |
ENSMUST00000027780.6
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr6_-_52181393 | 0.46 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr9_-_116004386 | 0.42 |
ENSMUST00000035014.8
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr8_+_106581719 | 0.40 |
ENSMUST00000040445.9
|
Thap11
|
THAP domain containing 11 |
| chr10_+_82534841 | 0.38 |
ENSMUST00000020478.14
|
Hcfc2
|
host cell factor C2 |
| chr7_-_103778992 | 0.37 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr8_+_79236051 | 0.27 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr17_+_5045178 | 0.20 |
ENSMUST00000092723.11
ENSMUST00000232180.2 ENSMUST00000115797.9 |
Arid1b
|
AT rich interactive domain 1B (SWI-like) |
| chr1_+_15875846 | 0.19 |
ENSMUST00000188371.7
ENSMUST00000027057.8 |
Terf1
|
telomeric repeat binding factor 1 |
| chr17_+_71511642 | 0.13 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr16_+_65612083 | 0.10 |
ENSMUST00000168064.3
|
Vgll3
|
vestigial like family member 3 |
| chr14_+_52860023 | 0.02 |
ENSMUST00000103570.2
|
Trav5-1
|
T cell receptor alpha variable 5-1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0006407 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 0.4 | 1.2 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 1.0 | GO:2000676 | fungiform papilla formation(GO:0061198) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 0.5 | GO:0060435 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.4 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 3.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.9 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.2 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 3.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.0 | 0.7 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 1.9 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 3.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 3.5 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 1.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 3.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 3.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 3.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 3.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |