Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
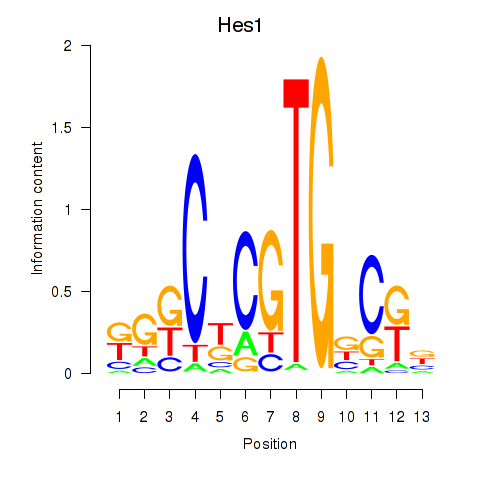
Results for Hes1
Z-value: 1.43
Transcription factors associated with Hes1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes1
|
ENSMUSG00000022528.9 | Hes1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes1 | mm39_v1_chr16_+_29884153_29884169 | 0.16 | 3.6e-01 | Click! |
Activity profile of Hes1 motif
Sorted Z-values of Hes1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hes1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_11976732 | 14.09 |
ENSMUST00000143915.2
|
Grb10
|
growth factor receptor bound protein 10 |
| chr11_-_11976237 | 11.27 |
ENSMUST00000150972.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr7_+_78563964 | 5.86 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr8_+_23629080 | 5.55 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr7_+_78564062 | 4.63 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr7_-_125968653 | 4.46 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr3_-_84386724 | 4.27 |
ENSMUST00000091002.8
|
Fhdc1
|
FH2 domain containing 1 |
| chr8_+_23629046 | 4.03 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr19_+_9995557 | 3.91 |
ENSMUST00000113161.10
ENSMUST00000238672.2 ENSMUST00000117641.8 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr11_-_102815910 | 3.61 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr8_+_95712151 | 3.52 |
ENSMUST00000212799.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr8_+_71858647 | 3.45 |
ENSMUST00000119976.8
ENSMUST00000120725.2 |
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chrX_+_70599524 | 3.33 |
ENSMUST00000072699.13
ENSMUST00000114582.9 ENSMUST00000015361.11 ENSMUST00000088874.10 |
Hmgb3
|
high mobility group box 3 |
| chr6_-_70769135 | 3.23 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr11_-_46203047 | 3.21 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr6_+_124806506 | 3.09 |
ENSMUST00000150120.8
|
Cdca3
|
cell division cycle associated 3 |
| chr5_+_123214332 | 3.03 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chrX_-_72974357 | 3.00 |
ENSMUST00000155597.2
ENSMUST00000114379.8 |
Renbp
|
renin binding protein |
| chr9_+_106306736 | 2.79 |
ENSMUST00000098994.7
ENSMUST00000059802.7 ENSMUST00000213448.2 ENSMUST00000217081.2 |
Rpl29
|
ribosomal protein L29 |
| chr17_+_25517363 | 2.77 |
ENSMUST00000037453.4
|
Prss34
|
protease, serine 34 |
| chr5_+_118165808 | 2.75 |
ENSMUST00000031304.14
|
Tesc
|
tescalcin |
| chr11_+_116089678 | 2.71 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chrX_-_72974440 | 2.70 |
ENSMUST00000116578.8
|
Renbp
|
renin binding protein |
| chr19_+_9995629 | 2.68 |
ENSMUST00000131407.2
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr11_+_116422712 | 2.64 |
ENSMUST00000100201.10
|
Sphk1
|
sphingosine kinase 1 |
| chr19_+_4644425 | 2.54 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr11_+_116422570 | 2.51 |
ENSMUST00000106387.9
|
Sphk1
|
sphingosine kinase 1 |
| chr5_+_139777263 | 2.50 |
ENSMUST00000018287.10
|
Mafk
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
| chr1_-_189420270 | 2.46 |
ENSMUST00000171929.8
ENSMUST00000165962.2 |
Cenpf
|
centromere protein F |
| chr4_+_130253925 | 2.43 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr5_-_36853281 | 2.41 |
ENSMUST00000031091.13
ENSMUST00000140653.2 |
D5Ertd579e
|
DNA segment, Chr 5, ERATO Doi 579, expressed |
| chr3_-_144466602 | 2.37 |
ENSMUST00000059091.6
|
Clca3a1
|
chloride channel accessory 3A1 |
| chr1_+_172327812 | 2.26 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr15_+_79575046 | 2.16 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr3_-_88410495 | 2.07 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr4_+_134195631 | 1.99 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr11_-_29497819 | 1.97 |
ENSMUST00000102844.4
|
Rps27a
|
ribosomal protein S27A |
| chr10_-_123032821 | 1.92 |
ENSMUST00000219619.2
ENSMUST00000020334.9 |
Usp15
|
ubiquitin specific peptidase 15 |
| chr10_+_11157326 | 1.88 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr8_-_123278054 | 1.88 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr6_-_31540913 | 1.86 |
ENSMUST00000026698.8
|
Podxl
|
podocalyxin-like |
| chr19_-_4356207 | 1.85 |
ENSMUST00000088737.11
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr7_-_126398343 | 1.81 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_126398165 | 1.79 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr12_-_36206626 | 1.74 |
ENSMUST00000220828.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr4_+_126915104 | 1.64 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr4_-_117740624 | 1.61 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr8_+_95393349 | 1.61 |
ENSMUST00000109527.6
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr13_-_74085880 | 1.55 |
ENSMUST00000022053.11
|
Trip13
|
thyroid hormone receptor interactor 13 |
| chr18_+_56840813 | 1.54 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1 |
| chr16_+_15705141 | 1.54 |
ENSMUST00000096232.6
|
Cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr5_+_122347792 | 1.54 |
ENSMUST00000072602.14
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr17_+_36172235 | 1.42 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr19_-_4355983 | 1.36 |
ENSMUST00000025791.12
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr3_+_54600196 | 1.35 |
ENSMUST00000197502.5
ENSMUST00000200441.5 ENSMUST00000029315.13 ENSMUST00000199655.5 |
Supt20
|
SPT20 SAGA complex component |
| chr14_-_57902411 | 1.31 |
ENSMUST00000089482.12
|
Xpo4
|
exportin 4 |
| chr14_-_51384236 | 1.28 |
ENSMUST00000080126.4
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr3_+_104545974 | 1.27 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr6_-_68681962 | 1.25 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr5_-_136199482 | 1.22 |
ENSMUST00000196454.5
ENSMUST00000197052.2 |
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr7_+_12758046 | 1.22 |
ENSMUST00000005705.8
|
Trim28
|
tripartite motif-containing 28 |
| chr2_+_127178072 | 1.21 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr5_+_122296322 | 1.19 |
ENSMUST00000102528.11
ENSMUST00000086294.11 |
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr11_-_45846291 | 1.18 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr3_-_102111892 | 1.17 |
ENSMUST00000029453.13
|
Vangl1
|
VANGL planar cell polarity 1 |
| chr3_+_54600509 | 1.14 |
ENSMUST00000170552.6
|
Supt20
|
SPT20 SAGA complex component |
| chr13_-_96803216 | 1.13 |
ENSMUST00000170287.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr11_-_106679983 | 1.12 |
ENSMUST00000129585.8
|
Ddx5
|
DEAD box helicase 5 |
| chr9_+_123902143 | 1.11 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr7_-_99132843 | 1.10 |
ENSMUST00000208532.2
ENSMUST00000107096.2 ENSMUST00000032998.13 |
Rps3
|
ribosomal protein S3 |
| chr17_+_24022153 | 1.09 |
ENSMUST00000190686.7
ENSMUST00000088621.11 ENSMUST00000233636.2 |
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr6_-_65121892 | 1.08 |
ENSMUST00000031982.5
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr3_+_37474422 | 1.08 |
ENSMUST00000029277.13
ENSMUST00000198968.2 |
Spata5
|
spermatogenesis associated 5 |
| chr15_+_84807640 | 1.07 |
ENSMUST00000230411.2
|
Nup50
|
nucleoporin 50 |
| chr18_-_60981981 | 1.06 |
ENSMUST00000177172.8
ENSMUST00000175934.8 ENSMUST00000176630.8 |
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr3_+_37694094 | 1.04 |
ENSMUST00000108109.8
ENSMUST00000038569.8 ENSMUST00000108107.2 |
Spry1
|
sprouty RTK signaling antagonist 1 |
| chr10_-_123032881 | 1.03 |
ENSMUST00000220377.2
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr4_-_129467430 | 1.03 |
ENSMUST00000102596.8
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr8_+_95393404 | 1.03 |
ENSMUST00000211858.2
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chrX_+_10351360 | 1.01 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr3_-_89959739 | 0.99 |
ENSMUST00000199929.2
ENSMUST00000090908.11 ENSMUST00000198322.5 ENSMUST00000196843.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr16_-_15964634 | 0.98 |
ENSMUST00000040248.9
|
Spidr
|
scaffolding protein involved in DNA repair |
| chr3_-_89959770 | 0.98 |
ENSMUST00000029553.16
ENSMUST00000195995.5 ENSMUST00000064639.15 ENSMUST00000199834.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr1_-_165462020 | 0.95 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chr2_-_144112700 | 0.94 |
ENSMUST00000110030.10
|
Snx5
|
sorting nexin 5 |
| chrX_-_105264751 | 0.94 |
ENSMUST00000113495.9
|
Taf9b
|
TATA-box binding protein associated factor 9B |
| chr11_+_43572825 | 0.94 |
ENSMUST00000061070.6
ENSMUST00000094294.5 |
Pwwp2a
|
PWWP domain containing 2A |
| chr2_-_84557694 | 0.94 |
ENSMUST00000028475.9
|
Clp1
|
CLP1, cleavage and polyadenylation factor I subunit |
| chr18_-_36648850 | 0.92 |
ENSMUST00000025363.7
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr15_-_76702170 | 0.92 |
ENSMUST00000175843.3
ENSMUST00000177026.3 ENSMUST00000176736.3 ENSMUST00000036176.16 ENSMUST00000176219.9 ENSMUST00000239134.2 ENSMUST00000239003.2 ENSMUST00000077821.10 |
Arhgap39
|
Rho GTPase activating protein 39 |
| chr2_-_84557580 | 0.91 |
ENSMUST00000165219.8
|
Clp1
|
CLP1, cleavage and polyadenylation factor I subunit |
| chr6_+_85408953 | 0.90 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr15_+_84807582 | 0.90 |
ENSMUST00000165443.4
|
Nup50
|
nucleoporin 50 |
| chr7_+_24584197 | 0.89 |
ENSMUST00000156372.8
ENSMUST00000124035.2 |
Rps19
|
ribosomal protein S19 |
| chr12_+_111504450 | 0.87 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr5_+_122347912 | 0.86 |
ENSMUST00000143560.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr16_-_90607251 | 0.85 |
ENSMUST00000140920.2
|
Urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr4_-_135080437 | 0.85 |
ENSMUST00000030613.11
ENSMUST00000131373.8 |
Srrm1
|
serine/arginine repetitive matrix 1 |
| chr4_+_108317197 | 0.85 |
ENSMUST00000097925.9
|
Tut4
|
terminal uridylyl transferase 4 |
| chr17_+_29251602 | 0.84 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr13_+_43519075 | 0.84 |
ENSMUST00000220458.2
ENSMUST00000220576.2 ENSMUST00000221515.2 |
Sirt5
|
sirtuin 5 |
| chr2_-_144112444 | 0.83 |
ENSMUST00000028909.5
|
Snx5
|
sorting nexin 5 |
| chr3_-_108819003 | 0.82 |
ENSMUST00000029480.9
|
Prpf38b
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B |
| chr3_-_57755500 | 0.82 |
ENSMUST00000066882.10
|
Pfn2
|
profilin 2 |
| chr14_-_30740946 | 0.82 |
ENSMUST00000228341.2
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr9_-_50528727 | 0.81 |
ENSMUST00000131351.8
ENSMUST00000171462.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr17_-_32583402 | 0.80 |
ENSMUST00000169488.10
|
Wiz
|
widely-interspaced zinc finger motifs |
| chr1_-_36312482 | 0.79 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr5_+_105563605 | 0.79 |
ENSMUST00000112707.3
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr11_+_68986043 | 0.78 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr11_-_106679671 | 0.78 |
ENSMUST00000123339.2
|
Ddx5
|
DEAD box helicase 5 |
| chr19_+_36811615 | 0.77 |
ENSMUST00000025729.12
|
Tnks2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr13_-_43457626 | 0.76 |
ENSMUST00000055341.7
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr2_+_120439858 | 0.75 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr11_-_32217547 | 0.75 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr19_+_47217279 | 0.75 |
ENSMUST00000111807.5
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr3_-_107993906 | 0.75 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr10_-_7831657 | 0.73 |
ENSMUST00000147938.2
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr8_-_107823141 | 0.72 |
ENSMUST00000068388.15
ENSMUST00000133925.8 ENSMUST00000068421.13 ENSMUST00000116425.3 |
Terf2
|
telomeric repeat binding factor 2 |
| chr5_+_108817133 | 0.72 |
ENSMUST00000119212.8
|
Idua
|
iduronidase, alpha-L |
| chr3_-_51248032 | 0.71 |
ENSMUST00000062009.14
ENSMUST00000194641.6 |
Elf2
|
E74-like factor 2 |
| chr2_+_153187729 | 0.70 |
ENSMUST00000227428.2
ENSMUST00000109790.2 |
Asxl1
|
ASXL transcriptional regulator 1 |
| chr9_+_111100893 | 0.70 |
ENSMUST00000135807.2
ENSMUST00000060711.8 |
Epm2aip1
|
EPM2A (laforin) interacting protein 1 |
| chr10_+_40225272 | 0.69 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr11_+_121325739 | 0.69 |
ENSMUST00000026175.9
ENSMUST00000092302.11 ENSMUST00000103014.4 |
Fn3k
|
fructosamine 3 kinase |
| chr11_+_101137786 | 0.67 |
ENSMUST00000107282.4
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr19_-_16758268 | 0.67 |
ENSMUST00000068156.8
|
Vps13a
|
vacuolar protein sorting 13A |
| chr8_+_95584146 | 0.66 |
ENSMUST00000211939.2
ENSMUST00000212124.2 |
Polr2c
|
polymerase (RNA) II (DNA directed) polypeptide C |
| chr8_+_83891972 | 0.65 |
ENSMUST00000034145.11
|
Tbc1d9
|
TBC1 domain family, member 9 |
| chr19_+_6952319 | 0.65 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr11_+_109253598 | 0.65 |
ENSMUST00000106702.4
ENSMUST00000020930.14 |
Gna13
|
guanine nucleotide binding protein, alpha 13 |
| chr14_+_25459630 | 0.65 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr17_-_48145466 | 0.65 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr13_-_34529157 | 0.64 |
ENSMUST00000040336.12
|
Slc22a23
|
solute carrier family 22, member 23 |
| chr5_+_74355940 | 0.64 |
ENSMUST00000051937.9
|
Rasl11b
|
RAS-like, family 11, member B |
| chr3_+_69224189 | 0.64 |
ENSMUST00000029355.9
|
Ppm1l
|
protein phosphatase 1 (formerly 2C)-like |
| chr15_-_38300937 | 0.64 |
ENSMUST00000227920.2
ENSMUST00000074043.7 ENSMUST00000228416.2 |
Klf10
|
Kruppel-like factor 10 |
| chr13_+_21364330 | 0.64 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr2_-_140012447 | 0.63 |
ENSMUST00000046030.8
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr4_+_108316568 | 0.63 |
ENSMUST00000106673.8
ENSMUST00000043368.12 |
Tut4
|
terminal uridylyl transferase 4 |
| chr3_+_28835425 | 0.62 |
ENSMUST00000060500.9
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr9_-_104140099 | 0.60 |
ENSMUST00000035170.13
|
Dnajc13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr17_+_83658354 | 0.59 |
ENSMUST00000096766.12
ENSMUST00000049503.10 ENSMUST00000112363.10 ENSMUST00000234460.2 |
Eml4
|
echinoderm microtubule associated protein like 4 |
| chr7_-_118455477 | 0.59 |
ENSMUST00000106550.11
|
Knop1
|
lysine rich nucleolar protein 1 |
| chr7_+_127475968 | 0.58 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr17_-_57338468 | 0.58 |
ENSMUST00000007814.10
ENSMUST00000233480.2 |
Khsrp
|
KH-type splicing regulatory protein |
| chr4_+_108704982 | 0.57 |
ENSMUST00000102738.4
|
Kti12
|
KTI12 homolog, chromatin associated |
| chr2_+_158636727 | 0.57 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr2_-_35869636 | 0.56 |
ENSMUST00000028248.11
ENSMUST00000112976.9 |
Ttll11
|
tubulin tyrosine ligase-like family, member 11 |
| chr9_-_50528530 | 0.54 |
ENSMUST00000147671.2
ENSMUST00000145139.8 ENSMUST00000155435.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr4_-_130068902 | 0.53 |
ENSMUST00000105998.8
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr11_-_116515026 | 0.52 |
ENSMUST00000103029.10
|
Rhbdf2
|
rhomboid 5 homolog 2 |
| chr14_-_104081119 | 0.52 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr19_+_30007910 | 0.52 |
ENSMUST00000025739.14
|
Uhrf2
|
ubiquitin-like, containing PHD and RING finger domains 2 |
| chr4_+_11191354 | 0.51 |
ENSMUST00000170901.8
|
Ccne2
|
cyclin E2 |
| chr1_-_175520185 | 0.48 |
ENSMUST00000104984.4
ENSMUST00000209720.2 ENSMUST00000211489.2 ENSMUST00000210367.2 ENSMUST00000027809.8 |
Chml
Opn3
|
choroideremia-like opsin 3 |
| chr3_+_54600268 | 0.47 |
ENSMUST00000199652.5
|
Supt20
|
SPT20 SAGA complex component |
| chr18_+_49965689 | 0.47 |
ENSMUST00000180611.8
|
Dmxl1
|
Dmx-like 1 |
| chr2_+_144210881 | 0.47 |
ENSMUST00000028911.15
ENSMUST00000147747.8 ENSMUST00000183618.3 |
Kat14
Pet117
|
lysine acetyltransferase 14 PET117 homolog |
| chr13_+_74085916 | 0.47 |
ENSMUST00000222399.2
ENSMUST00000099384.4 |
Brd9
|
bromodomain containing 9 |
| chr1_-_143578542 | 0.47 |
ENSMUST00000018337.9
|
Cdc73
|
cell division cycle 73, Paf1/RNA polymerase II complex component |
| chr5_-_65855199 | 0.47 |
ENSMUST00000031104.7
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr11_-_115977755 | 0.46 |
ENSMUST00000074628.13
ENSMUST00000106444.4 |
Wbp2
|
WW domain binding protein 2 |
| chr5_+_135178509 | 0.46 |
ENSMUST00000153183.8
|
Tbl2
|
transducin (beta)-like 2 |
| chr5_+_64316771 | 0.46 |
ENSMUST00000043893.13
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr4_-_135080471 | 0.44 |
ENSMUST00000084846.12
ENSMUST00000136342.9 ENSMUST00000105861.8 |
Srrm1
|
serine/arginine repetitive matrix 1 |
| chr11_-_45845992 | 0.44 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr3_-_89959917 | 0.44 |
ENSMUST00000197903.5
|
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr11_+_106680062 | 0.42 |
ENSMUST00000103068.10
ENSMUST00000018516.11 |
Cep95
|
centrosomal protein 95 |
| chr4_-_129590609 | 0.42 |
ENSMUST00000102588.10
|
Tmem39b
|
transmembrane protein 39b |
| chr12_-_101924407 | 0.41 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr8_-_70963202 | 0.41 |
ENSMUST00000125184.8
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr11_-_97877219 | 0.41 |
ENSMUST00000107565.3
ENSMUST00000107564.2 ENSMUST00000017561.15 |
Plxdc1
|
plexin domain containing 1 |
| chr7_+_66872321 | 0.40 |
ENSMUST00000207757.2
ENSMUST00000058771.13 ENSMUST00000207823.2 ENSMUST00000208698.2 ENSMUST00000208998.2 ENSMUST00000179106.3 |
Lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr18_+_23937019 | 0.39 |
ENSMUST00000025127.5
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr16_-_89940652 | 0.39 |
ENSMUST00000114124.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr13_-_23735822 | 0.39 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr18_-_80756273 | 0.39 |
ENSMUST00000078049.12
ENSMUST00000236711.2 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr16_+_93680846 | 0.39 |
ENSMUST00000142316.2
|
Chaf1b
|
chromatin assembly factor 1, subunit B (p60) |
| chr9_+_56979307 | 0.38 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr15_+_84565174 | 0.37 |
ENSMUST00000065499.5
|
Prr5
|
proline rich 5 (renal) |
| chr9_-_50650663 | 0.37 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr6_-_12749192 | 0.37 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr12_+_33479604 | 0.35 |
ENSMUST00000020877.9
|
Polr1f
|
RNA polymerase I subunit F |
| chr10_+_22520910 | 0.34 |
ENSMUST00000042261.5
|
Slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr17_-_36582721 | 0.33 |
ENSMUST00000042717.13
|
Trim39
|
tripartite motif-containing 39 |
| chr1_+_58432629 | 0.33 |
ENSMUST00000186949.2
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr18_+_67476664 | 0.32 |
ENSMUST00000025404.10
|
Cidea
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| chr11_+_22462088 | 0.32 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr12_-_114012399 | 0.31 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr2_+_157870606 | 0.31 |
ENSMUST00000109518.8
ENSMUST00000029180.14 |
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chrX_+_41156713 | 0.31 |
ENSMUST00000115094.8
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr4_-_144973423 | 0.30 |
ENSMUST00000030336.11
|
Tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1b |
| chr19_-_6593049 | 0.29 |
ENSMUST00000113451.9
|
Slc22a12
|
solute carrier family 22 (organic anion/cation transporter), member 12 |
| chr8_-_70962972 | 0.29 |
ENSMUST00000140679.8
ENSMUST00000129909.8 ENSMUST00000081940.11 |
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr16_+_32238520 | 0.29 |
ENSMUST00000014220.15
ENSMUST00000080316.8 |
Dynlt2b
|
dynein light chain Tctex-type 2B |
| chr4_+_110254907 | 0.28 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr4_+_156028281 | 0.28 |
ENSMUST00000103175.8
ENSMUST00000166489.8 ENSMUST00000024056.10 ENSMUST00000136492.2 ENSMUST00000105583.9 ENSMUST00000152536.8 ENSMUST00000118192.8 |
Ube2j2
|
ubiquitin-conjugating enzyme E2J 2 |
| chr11_-_59678462 | 0.28 |
ENSMUST00000125307.2
|
Pld6
|
phospholipase D family, member 6 |
| chr2_+_152135748 | 0.28 |
ENSMUST00000028963.14
ENSMUST00000144252.8 |
Tbc1d20
|
TBC1 domain family, member 20 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 25.4 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 1.7 | 10.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 5.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.8 | 3.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 5.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.7 | 4.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.7 | 2.7 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.6 | 3.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.6 | 3.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.5 | 2.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 | 1.2 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.4 | 3.2 | GO:0042699 | tachykinin receptor signaling pathway(GO:0007217) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.4 | 1.5 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.4 | 3.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.4 | 1.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 1.1 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.3 | 2.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 3.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 1.8 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.3 | 0.9 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.3 | 2.0 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 1.1 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.3 | 4.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.0 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.2 | 2.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 2.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.9 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.2 | 3.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 8.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 0.7 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 1.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 | 1.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 2.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.2 | 1.9 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.7 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 1.9 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.6 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.4 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.1 | 0.4 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.8 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 2.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 0.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 2.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.6 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.3 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 3.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 1.0 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 0.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.3 | GO:0007315 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.1 | 0.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.5 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.6 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 2.7 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.1 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 1.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 1.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 3.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 2.5 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 1.1 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.8 | GO:1904814 | regulation of protein localization to chromosome, telomeric region(GO:1904814) positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 1.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 1.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.8 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.5 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.0 | 0.2 | GO:1903719 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 2.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.0 | GO:0060003 | copper ion export(GO:0060003) |
| 0.0 | 0.3 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.7 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 1.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.7 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 0.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 1.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 6.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.9 | GO:0051291 | protein heterooligomerization(GO:0051291) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.5 | 3.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 3.6 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.3 | 3.5 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.3 | 1.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 8.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.3 | 0.5 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 3.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 1.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 2.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 10.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 3.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 1.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 2.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 2.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 3.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 3.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 4.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 4.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 3.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 3.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.1 | 3.2 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.9 | 8.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.6 | 2.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.6 | 1.9 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.6 | 3.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.6 | 5.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.5 | 1.6 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 1.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 29.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.4 | 1.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.4 | 2.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.4 | 1.1 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.4 | 1.1 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.4 | 1.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.4 | 3.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 5.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 3.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 1.8 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.3 | 2.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.3 | 1.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 1.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.8 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.2 | 0.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.0 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 1.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.3 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 1.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 6.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.4 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 3.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 3.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 2.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 3.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 3.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 3.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 25.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 4.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 5.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 4.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 3.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 25.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 3.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 8.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 5.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 10.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 5.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 3.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 3.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 2.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 4.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 2.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |