Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
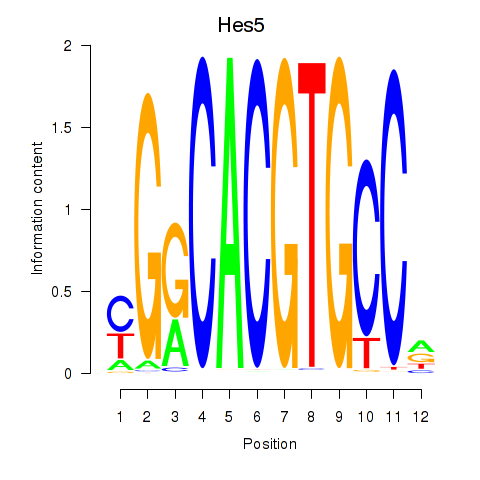
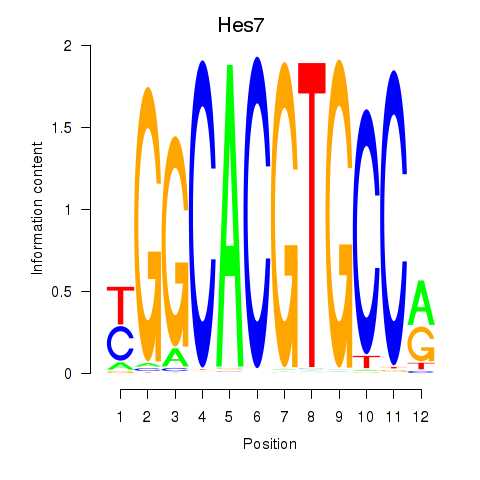
Results for Hes5_Hes7
Z-value: 1.75
Transcription factors associated with Hes5_Hes7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes5
|
ENSMUSG00000048001.8 | Hes5 |
|
Hes7
|
ENSMUSG00000023781.3 | Hes7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes7 | mm39_v1_chr11_+_69011230_69011230 | 0.65 | 1.8e-05 | Click! |
| Hes5 | mm39_v1_chr4_+_155045372_155045387 | 0.44 | 7.0e-03 | Click! |
Activity profile of Hes5_Hes7 motif
Sorted Z-values of Hes5_Hes7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hes5_Hes7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_8137372 | 17.27 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_+_8137620 | 16.30 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr11_-_11976732 | 15.88 |
ENSMUST00000143915.2
|
Grb10
|
growth factor receptor bound protein 10 |
| chrX_+_8137881 | 15.07 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr11_-_11976237 | 12.73 |
ENSMUST00000150972.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr17_-_26417982 | 12.15 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr13_+_108452866 | 10.67 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B |
| chr13_+_108452930 | 7.53 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B |
| chr4_+_134195631 | 7.48 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr9_-_21874802 | 6.49 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr12_+_109425769 | 5.93 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr2_+_150412329 | 5.64 |
ENSMUST00000089200.3
|
Cst7
|
cystatin F (leukocystatin) |
| chr15_-_78739717 | 4.94 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr6_+_72074718 | 3.98 |
ENSMUST00000187007.3
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr5_+_31409021 | 3.59 |
ENSMUST00000054829.13
ENSMUST00000201625.4 ENSMUST00000201937.4 |
Krtcap3
|
keratinocyte associated protein 3 |
| chr6_+_72074545 | 3.55 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr9_+_107464841 | 3.53 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr17_-_25493273 | 3.05 |
ENSMUST00000172587.8
ENSMUST00000049911.16 ENSMUST00000173713.8 |
Ube2i
|
ubiquitin-conjugating enzyme E2I |
| chr1_+_172327812 | 2.89 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr17_-_15181491 | 2.85 |
ENSMUST00000024657.12
|
Phf10
|
PHD finger protein 10 |
| chr6_+_35154545 | 2.69 |
ENSMUST00000170234.2
|
Nup205
|
nucleoporin 205 |
| chr3_-_95763065 | 2.50 |
ENSMUST00000161476.8
|
Prpf3
|
pre-mRNA processing factor 3 |
| chr9_-_107167046 | 2.34 |
ENSMUST00000035194.8
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr3_-_95763160 | 2.34 |
ENSMUST00000015892.14
|
Prpf3
|
pre-mRNA processing factor 3 |
| chr17_-_25492832 | 2.32 |
ENSMUST00000172868.8
ENSMUST00000172618.8 |
Ube2i
|
ubiquitin-conjugating enzyme E2I |
| chr4_+_154954042 | 2.05 |
ENSMUST00000079269.14
ENSMUST00000163732.8 ENSMUST00000080559.13 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr17_+_48623157 | 1.85 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr17_+_36172210 | 1.83 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr11_+_3913970 | 1.81 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr1_-_192718064 | 1.74 |
ENSMUST00000215093.2
ENSMUST00000195354.6 |
Syt14
|
synaptotagmin XIV |
| chrX_-_9123138 | 1.68 |
ENSMUST00000115553.3
|
Gm14862
|
predicted gene 14862 |
| chr11_+_101207021 | 1.66 |
ENSMUST00000142640.8
ENSMUST00000019470.14 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr10_+_85665234 | 1.56 |
ENSMUST00000217667.2
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr2_-_92201342 | 1.33 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr1_-_188740023 | 1.10 |
ENSMUST00000085678.8
|
Kctd3
|
potassium channel tetramerisation domain containing 3 |
| chr1_-_161704224 | 1.10 |
ENSMUST00000048377.11
|
Suco
|
SUN domain containing ossification factor |
| chr4_-_43562397 | 1.07 |
ENSMUST00000030187.14
|
Tln1
|
talin 1 |
| chr4_+_32657105 | 1.06 |
ENSMUST00000071642.11
ENSMUST00000178134.2 |
Mdn1
|
midasin AAA ATPase 1 |
| chr19_+_47167259 | 1.05 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr9_-_36637923 | 1.00 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr15_-_98507913 | 0.99 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr17_+_28945057 | 0.98 |
ENSMUST00000233003.2
|
Gm4356
|
predicted gene 4356 |
| chr2_+_91357100 | 0.93 |
ENSMUST00000111338.10
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr1_-_192717958 | 0.93 |
ENSMUST00000016344.9
|
Syt14
|
synaptotagmin XIV |
| chr1_-_75195127 | 0.87 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr1_-_75195889 | 0.80 |
ENSMUST00000186213.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr9_+_20563386 | 0.79 |
ENSMUST00000034689.8
|
Pin1
|
protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1 |
| chr9_-_36637670 | 0.70 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr18_+_37453427 | 0.70 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr8_-_25438784 | 0.66 |
ENSMUST00000119720.8
ENSMUST00000121438.9 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr11_+_79883885 | 0.64 |
ENSMUST00000163272.2
ENSMUST00000017692.15 |
Suz12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr14_-_30740946 | 0.63 |
ENSMUST00000228341.2
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr7_-_130964469 | 0.62 |
ENSMUST00000059438.11
|
2310057M21Rik
|
RIKEN cDNA 2310057M21 gene |
| chr6_-_52203146 | 0.58 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr18_-_38131766 | 0.56 |
ENSMUST00000236588.2
ENSMUST00000237272.2 ENSMUST00000236134.2 |
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr6_+_127538268 | 0.52 |
ENSMUST00000212051.2
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr3_+_104545974 | 0.51 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr4_-_132484559 | 0.51 |
ENSMUST00000030709.9
|
Smpdl3b
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr14_-_69945022 | 0.48 |
ENSMUST00000118374.8
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr18_-_35855383 | 0.47 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr4_-_133484080 | 0.46 |
ENSMUST00000008024.7
|
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr10_+_36850532 | 0.42 |
ENSMUST00000019911.14
ENSMUST00000105510.2 |
Hdac2
|
histone deacetylase 2 |
| chr11_+_58197881 | 0.40 |
ENSMUST00000153510.9
|
Zfp692
|
zinc finger protein 692 |
| chr14_-_69944942 | 0.39 |
ENSMUST00000121142.4
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr15_-_37004302 | 0.37 |
ENSMUST00000228275.2
|
Zfp706
|
zinc finger protein 706 |
| chr8_+_125448867 | 0.35 |
ENSMUST00000034463.4
|
Arv1
|
ARV1 homolog, fatty acid homeostasis modulator |
| chr16_+_45044678 | 0.34 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr19_+_8713156 | 0.32 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr1_-_131161312 | 0.29 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chrX_-_8042129 | 0.28 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr9_-_78350486 | 0.26 |
ENSMUST00000070742.14
ENSMUST00000034898.14 |
Cgas
|
cyclic GMP-AMP synthase |
| chr2_-_92201311 | 0.25 |
ENSMUST00000176774.8
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr10_-_57408512 | 0.24 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr11_+_54757200 | 0.24 |
ENSMUST00000020504.6
|
Hint1
|
histidine triad nucleotide binding protein 1 |
| chr8_+_13155621 | 0.22 |
ENSMUST00000016680.14
ENSMUST00000121426.2 |
Cul4a
|
cullin 4A |
| chr11_+_4587733 | 0.19 |
ENSMUST00000070257.14
ENSMUST00000109930.3 |
Ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr9_+_75221415 | 0.18 |
ENSMUST00000215875.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr6_-_145021816 | 0.18 |
ENSMUST00000111742.8
ENSMUST00000048252.11 |
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr15_+_101982208 | 0.14 |
ENSMUST00000169681.3
ENSMUST00000229400.2 |
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr16_-_33787399 | 0.14 |
ENSMUST00000023510.7
|
Umps
|
uridine monophosphate synthetase |
| chr11_-_59678462 | 0.14 |
ENSMUST00000125307.2
|
Pld6
|
phospholipase D family, member 6 |
| chr11_+_22462088 | 0.14 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr10_+_79518002 | 0.14 |
ENSMUST00000020550.13
|
Cdc34
|
cell division cycle 34 |
| chr12_+_77285770 | 0.11 |
ENSMUST00000062804.8
|
Fut8
|
fucosyltransferase 8 |
| chr4_+_123798690 | 0.10 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr10_-_80223475 | 0.10 |
ENSMUST00000105350.3
|
Mex3d
|
mex3 RNA binding family member D |
| chr6_+_71520855 | 0.09 |
ENSMUST00000204535.2
ENSMUST00000065364.5 ENSMUST00000204199.2 |
Chmp3
|
charged multivesicular body protein 3 |
| chr15_+_38662158 | 0.07 |
ENSMUST00000022904.8
ENSMUST00000228820.2 |
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chr3_-_95148909 | 0.06 |
ENSMUST00000090815.6
ENSMUST00000107197.2 |
Gm128
|
predicted gene 128 |
| chr9_+_64028783 | 0.06 |
ENSMUST00000118215.3
|
Lctl
|
lactase-like |
| chr4_+_123798625 | 0.04 |
ENSMUST00000030400.14
|
Mycbp
|
MYC binding protein |
| chr5_+_122788469 | 0.02 |
ENSMUST00000199371.2
|
P2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr5_+_43672856 | 0.01 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr10_+_79518138 | 0.00 |
ENSMUST00000166603.2
ENSMUST00000219791.2 ENSMUST00000219930.2 ENSMUST00000218964.2 |
Cdc34
|
cell division cycle 34 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 28.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 2.7 | 48.7 | GO:0015816 | glycine transport(GO:0015816) |
| 1.1 | 7.5 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.9 | 5.4 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.6 | 1.7 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.4 | 6.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.4 | 4.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 1.0 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.3 | 1.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.2 | 10.5 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.2 | 2.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 0.9 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 5.9 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 | 2.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) nucleolus organization(GO:0007000) |
| 0.1 | 1.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 7.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 18.5 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.4 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0007315 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.5 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 5.2 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 1.1 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.6 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.1 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 2.7 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 1.7 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.4 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.4 | 2.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 1.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 4.8 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 3.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 44.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 7.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 8.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 48.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 2.5 | 7.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.0 | 12.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.9 | 5.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.5 | 4.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 28.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.4 | 1.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 1.0 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.2 | 1.6 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 2.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 1.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.4 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 2.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 5.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 6.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 14.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 7.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 28.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 5.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 12.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 6.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 28.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.7 | 48.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 5.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 5.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 18.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |