Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hic1
Z-value: 2.45
Transcription factors associated with Hic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hic1
|
ENSMUSG00000043099.5 | Hic1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hic1 | mm39_v1_chr11_-_75060345_75060345 | -0.27 | 1.1e-01 | Click! |
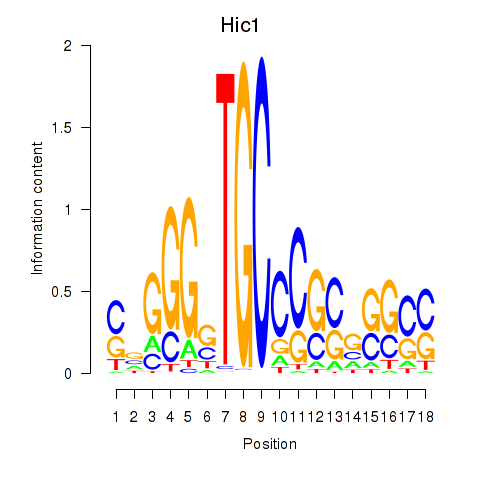
Activity profile of Hic1 motif
Sorted Z-values of Hic1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hic1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_94102255 | 9.23 |
ENSMUST00000041589.6
|
Tob1
|
transducer of ErbB-2.1 |
| chr8_-_85526653 | 8.71 |
ENSMUST00000126806.2
ENSMUST00000076715.13 |
Nfix
|
nuclear factor I/X |
| chr2_-_5719302 | 8.24 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr5_-_110434026 | 6.81 |
ENSMUST00000031472.12
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chr4_+_141095415 | 6.63 |
ENSMUST00000006380.5
|
Fam131c
|
family with sequence similarity 131, member C |
| chr9_-_42175522 | 6.60 |
ENSMUST00000217513.2
ENSMUST00000052725.15 |
Sc5d
|
sterol-C5-desaturase |
| chr7_+_65511777 | 6.56 |
ENSMUST00000055576.12
ENSMUST00000098391.11 |
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr8_+_96078886 | 6.42 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chr9_-_42175496 | 6.35 |
ENSMUST00000169609.2
|
Sc5d
|
sterol-C5-desaturase |
| chr3_-_121608809 | 6.18 |
ENSMUST00000197383.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr3_-_18297451 | 5.88 |
ENSMUST00000035625.7
|
Cyp7b1
|
cytochrome P450, family 7, subfamily b, polypeptide 1 |
| chr16_-_45830575 | 5.88 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr8_+_127790772 | 5.85 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr3_-_121608859 | 5.73 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr16_+_26400454 | 5.44 |
ENSMUST00000096129.9
ENSMUST00000166294.9 ENSMUST00000174202.8 ENSMUST00000023156.13 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr18_+_51250748 | 5.32 |
ENSMUST00000116639.4
|
Prr16
|
proline rich 16 |
| chr7_-_114162125 | 5.21 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr15_+_7159038 | 5.17 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr19_+_57599452 | 5.16 |
ENSMUST00000077282.7
|
Atrnl1
|
attractin like 1 |
| chr1_+_72863641 | 5.13 |
ENSMUST00000047328.11
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr7_+_65511482 | 5.12 |
ENSMUST00000176199.8
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr18_+_21094477 | 5.11 |
ENSMUST00000234316.2
|
Rnf125
|
ring finger protein 125 |
| chr3_+_40905066 | 5.09 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr12_-_75782502 | 5.08 |
ENSMUST00000021450.6
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr17_+_13980764 | 4.87 |
ENSMUST00000139347.8
ENSMUST00000156591.8 ENSMUST00000170827.9 ENSMUST00000139666.8 ENSMUST00000137708.8 ENSMUST00000137784.8 ENSMUST00000150848.8 |
Afdn
|
afadin, adherens junction formation factor |
| chr10_-_83173708 | 4.80 |
ENSMUST00000039956.6
|
Slc41a2
|
solute carrier family 41, member 2 |
| chr13_-_69759541 | 4.78 |
ENSMUST00000091514.6
|
Srd5a1
|
steroid 5 alpha-reductase 1 |
| chr4_-_46991842 | 4.78 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr6_+_17463925 | 4.71 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr19_+_44977512 | 4.68 |
ENSMUST00000026225.15
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr9_-_66882669 | 4.61 |
ENSMUST00000215172.2
ENSMUST00000034929.7 |
Lactb
|
lactamase, beta |
| chr1_+_182591425 | 4.56 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr18_-_60881679 | 4.40 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr2_-_65068960 | 4.35 |
ENSMUST00000112429.9
ENSMUST00000102726.8 ENSMUST00000112430.8 |
Cobll1
|
Cobl-like 1 |
| chr7_+_114014509 | 4.28 |
ENSMUST00000032909.9
|
Pde3b
|
phosphodiesterase 3B, cGMP-inhibited |
| chr11_-_87878301 | 4.24 |
ENSMUST00000020775.9
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr14_-_55983142 | 4.18 |
ENSMUST00000002403.10
|
Dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr9_+_121471782 | 4.16 |
ENSMUST00000035115.5
|
Vipr1
|
vasoactive intestinal peptide receptor 1 |
| chr2_+_30254239 | 4.07 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chr19_-_29025233 | 4.03 |
ENSMUST00000025696.5
|
Ak3
|
adenylate kinase 3 |
| chr8_-_85526972 | 3.97 |
ENSMUST00000099070.10
|
Nfix
|
nuclear factor I/X |
| chr2_+_24944367 | 3.95 |
ENSMUST00000100334.11
ENSMUST00000152122.8 ENSMUST00000116574.10 ENSMUST00000006646.15 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr12_+_78273144 | 3.86 |
ENSMUST00000052472.6
|
Gphn
|
gephyrin |
| chr2_+_24944407 | 3.85 |
ENSMUST00000102931.11
ENSMUST00000074422.14 ENSMUST00000132172.8 ENSMUST00000114388.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr2_+_76505619 | 3.81 |
ENSMUST00000111920.2
|
Plekha3
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 3 |
| chr16_+_38722666 | 3.80 |
ENSMUST00000023478.8
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr16_+_31482658 | 3.80 |
ENSMUST00000115201.8
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr3_+_40904253 | 3.79 |
ENSMUST00000048490.13
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr10_-_125225298 | 3.77 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr13_-_64277115 | 3.74 |
ENSMUST00000220792.2
ENSMUST00000222866.2 ENSMUST00000099441.6 ENSMUST00000222168.2 |
Slc35d2
|
solute carrier family 35, member D2 |
| chr4_-_104967032 | 3.70 |
ENSMUST00000030243.8
|
Prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr18_-_60881405 | 3.69 |
ENSMUST00000237070.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr14_+_21126204 | 3.68 |
ENSMUST00000224069.2
|
Adk
|
adenosine kinase |
| chr6_+_116241146 | 3.58 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr5_-_31453206 | 3.58 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr2_+_102489558 | 3.53 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr16_+_24212284 | 3.49 |
ENSMUST00000038053.14
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr4_-_57143437 | 3.42 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr17_-_31855605 | 3.42 |
ENSMUST00000151718.3
ENSMUST00000135425.9 ENSMUST00000155814.8 |
Cbs
|
cystathionine beta-synthase |
| chr19_+_3373285 | 3.40 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr14_-_34032450 | 3.38 |
ENSMUST00000227375.2
|
Shld2
|
shieldin complex subunit 2 |
| chr16_+_31482745 | 3.36 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr4_+_138181616 | 3.30 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr4_+_40722911 | 3.29 |
ENSMUST00000164233.8
ENSMUST00000137246.8 ENSMUST00000125442.8 |
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr11_+_48728291 | 3.27 |
ENSMUST00000046903.6
|
Trim7
|
tripartite motif-containing 7 |
| chr8_+_56747613 | 3.21 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr2_+_155223728 | 3.21 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr2_-_65068917 | 3.18 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr10_-_31485180 | 3.15 |
ENSMUST00000081989.8
|
Rnf217
|
ring finger protein 217 |
| chr6_+_17463748 | 3.14 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr13_+_74787952 | 3.14 |
ENSMUST00000221822.2
ENSMUST00000221526.2 |
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr17_-_31856109 | 3.12 |
ENSMUST00000078509.12
ENSMUST00000236853.2 ENSMUST00000067801.14 ENSMUST00000118504.9 |
Cbs
|
cystathionine beta-synthase |
| chr2_+_24944480 | 3.10 |
ENSMUST00000114386.8
|
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr2_-_65069065 | 3.07 |
ENSMUST00000112431.8
|
Cobll1
|
Cobl-like 1 |
| chr5_-_116560916 | 3.06 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr16_+_31482949 | 3.05 |
ENSMUST00000023454.12
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr8_-_84059048 | 3.04 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr1_+_87254729 | 3.04 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chrX_+_70408351 | 3.03 |
ENSMUST00000146213.8
ENSMUST00000114601.8 ENSMUST00000015358.8 |
Mtmr1
|
myotubularin related protein 1 |
| chr15_-_31531122 | 3.02 |
ENSMUST00000090227.6
|
Marchf6
|
membrane associated ring-CH-type finger 6 |
| chr13_-_104246084 | 3.00 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr5_-_34345014 | 2.99 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr1_-_10038030 | 2.97 |
ENSMUST00000185184.2
|
Tcf24
|
transcription factor 24 |
| chr19_-_41252370 | 2.97 |
ENSMUST00000237871.2
ENSMUST00000025989.10 |
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr12_+_78273356 | 2.93 |
ENSMUST00000110388.10
|
Gphn
|
gephyrin |
| chr18_+_84106796 | 2.93 |
ENSMUST00000235383.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr2_+_127112613 | 2.93 |
ENSMUST00000125049.2
ENSMUST00000110374.2 |
Stard7
|
START domain containing 7 |
| chr2_+_102488985 | 2.89 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_-_65069383 | 2.86 |
ENSMUST00000155916.8
ENSMUST00000156643.2 |
Cobll1
|
Cobl-like 1 |
| chr5_+_105848598 | 2.85 |
ENSMUST00000120847.8
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr17_+_26934617 | 2.84 |
ENSMUST00000062519.14
ENSMUST00000144221.2 ENSMUST00000142539.8 ENSMUST00000151681.2 |
Crebrf
|
CREB3 regulatory factor |
| chr9_+_47441471 | 2.84 |
ENSMUST00000114548.8
ENSMUST00000152459.8 ENSMUST00000143026.9 ENSMUST00000085909.9 ENSMUST00000114547.8 ENSMUST00000239368.2 ENSMUST00000214542.2 ENSMUST00000034581.4 |
Cadm1
|
cell adhesion molecule 1 |
| chr1_-_132067343 | 2.81 |
ENSMUST00000112362.3
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr2_-_165997179 | 2.79 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr6_+_90527762 | 2.78 |
ENSMUST00000130418.8
ENSMUST00000032175.11 ENSMUST00000203111.2 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr18_-_62044871 | 2.77 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr19_+_32734884 | 2.76 |
ENSMUST00000013807.8
|
Pten
|
phosphatase and tensin homolog |
| chr10_-_81262948 | 2.73 |
ENSMUST00000078185.14
ENSMUST00000020461.15 ENSMUST00000105321.10 |
Nfic
|
nuclear factor I/C |
| chrX_-_50106844 | 2.71 |
ENSMUST00000053593.8
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr19_+_7034149 | 2.70 |
ENSMUST00000040261.7
|
Macrod1
|
mono-ADP ribosylhydrolase 1 |
| chr2_+_52747855 | 2.69 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr17_+_79922329 | 2.69 |
ENSMUST00000040368.3
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr7_+_45354512 | 2.68 |
ENSMUST00000080885.12
ENSMUST00000211513.2 ENSMUST00000211357.2 |
Dbp
|
D site albumin promoter binding protein |
| chr16_-_38342949 | 2.68 |
ENSMUST00000002925.6
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr2_+_155224105 | 2.68 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr11_-_61745843 | 2.67 |
ENSMUST00000004920.4
|
Ulk2
|
unc-51 like kinase 2 |
| chr8_-_71834543 | 2.64 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr8_+_124380639 | 2.64 |
ENSMUST00000045487.4
|
Rhou
|
ras homolog family member U |
| chr6_+_91661034 | 2.64 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr9_-_110571645 | 2.63 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr19_+_37423198 | 2.63 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr17_+_64170967 | 2.63 |
ENSMUST00000232945.2
|
Fer
|
fer (fms/fps related) protein kinase |
| chr5_-_33940053 | 2.62 |
ENSMUST00000148451.2
ENSMUST00000005431.6 |
Letm1
|
leucine zipper-EF-hand containing transmembrane protein 1 |
| chr3_-_116762617 | 2.61 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr17_+_79922486 | 2.61 |
ENSMUST00000225357.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr16_-_30207348 | 2.60 |
ENSMUST00000061350.13
ENSMUST00000100013.9 |
Atp13a3
|
ATPase type 13A3 |
| chr9_+_14187597 | 2.58 |
ENSMUST00000208222.2
|
Sesn3
|
sestrin 3 |
| chr10_-_78188301 | 2.57 |
ENSMUST00000138035.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr7_-_30672747 | 2.55 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr7_+_44114815 | 2.54 |
ENSMUST00000035929.11
ENSMUST00000146128.8 |
Aspdh
|
aspartate dehydrogenase domain containing |
| chr18_+_64473091 | 2.53 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr7_-_30672824 | 2.53 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr6_+_115337899 | 2.53 |
ENSMUST00000171644.8
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr2_+_116951855 | 2.53 |
ENSMUST00000028829.13
|
Spred1
|
sprouty protein with EVH-1 domain 1, related sequence |
| chr7_-_67022520 | 2.52 |
ENSMUST00000156690.8
ENSMUST00000107476.8 ENSMUST00000076325.12 ENSMUST00000032776.15 ENSMUST00000133074.2 |
Mef2a
|
myocyte enhancer factor 2A |
| chr1_+_16175998 | 2.51 |
ENSMUST00000027053.8
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr8_-_94738748 | 2.50 |
ENSMUST00000143265.2
|
Amfr
|
autocrine motility factor receptor |
| chr15_+_25843225 | 2.50 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr8_-_123884968 | 2.50 |
ENSMUST00000137998.2
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr5_-_62923463 | 2.50 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_-_30672889 | 2.49 |
ENSMUST00000001279.15
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr5_+_73648368 | 2.49 |
ENSMUST00000113558.8
ENSMUST00000063882.12 |
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr13_+_38010879 | 2.49 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr2_+_132105056 | 2.48 |
ENSMUST00000110158.8
ENSMUST00000103181.11 |
Cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr7_+_44114857 | 2.46 |
ENSMUST00000135624.2
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr5_-_76452577 | 2.45 |
ENSMUST00000202651.4
|
Clock
|
circadian locomotor output cycles kaput |
| chr19_-_44058175 | 2.45 |
ENSMUST00000172041.8
ENSMUST00000071698.13 ENSMUST00000112028.10 |
Erlin1
|
ER lipid raft associated 1 |
| chr9_-_107586678 | 2.44 |
ENSMUST00000193108.6
|
Sema3f
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr9_-_105372235 | 2.44 |
ENSMUST00000176190.8
ENSMUST00000163879.9 ENSMUST00000112558.10 ENSMUST00000176363.9 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr4_-_103071988 | 2.43 |
ENSMUST00000036195.13
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr6_-_43643093 | 2.43 |
ENSMUST00000114644.8
ENSMUST00000067888.14 |
Tpk1
|
thiamine pyrophosphokinase |
| chr2_-_104573179 | 2.42 |
ENSMUST00000028595.8
|
Depdc7
|
DEP domain containing 7 |
| chr1_+_182591771 | 2.42 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr14_+_45588509 | 2.42 |
ENSMUST00000226873.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chr15_-_89064936 | 2.42 |
ENSMUST00000109331.9
|
Plxnb2
|
plexin B2 |
| chr7_-_81356653 | 2.42 |
ENSMUST00000026922.15
|
Homer2
|
homer scaffolding protein 2 |
| chr7_-_81356557 | 2.42 |
ENSMUST00000207983.2
|
Homer2
|
homer scaffolding protein 2 |
| chr11_+_98239230 | 2.39 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr11_-_23583591 | 2.39 |
ENSMUST00000180260.8
ENSMUST00000141353.8 ENSMUST00000131612.2 ENSMUST00000109532.9 |
0610010F05Rik
|
RIKEN cDNA 0610010F05 gene |
| chr2_-_131987008 | 2.38 |
ENSMUST00000028815.15
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr11_+_84070593 | 2.38 |
ENSMUST00000137500.9
ENSMUST00000130012.9 |
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr19_-_44057800 | 2.37 |
ENSMUST00000170801.8
|
Erlin1
|
ER lipid raft associated 1 |
| chr15_+_90108480 | 2.36 |
ENSMUST00000100309.3
ENSMUST00000231200.2 |
Alg10b
|
asparagine-linked glycosylation 10B (alpha-1,2-glucosyltransferase) |
| chr1_+_131898325 | 2.34 |
ENSMUST00000027695.8
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr7_-_34914675 | 2.33 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr10_-_127456791 | 2.29 |
ENSMUST00000118455.2
ENSMUST00000121829.8 |
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr18_-_16942289 | 2.28 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr5_-_122639840 | 2.28 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr19_-_34856853 | 2.27 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr2_+_4722956 | 2.26 |
ENSMUST00000056914.7
|
Bend7
|
BEN domain containing 7 |
| chr2_+_127112127 | 2.24 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr5_+_105847811 | 2.24 |
ENSMUST00000060531.16
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr7_-_81104423 | 2.23 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr4_-_81360993 | 2.23 |
ENSMUST00000107262.8
ENSMUST00000102830.10 |
Mpdz
|
multiple PDZ domain crumbs cell polarity complex component |
| chr6_+_17463819 | 2.23 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr3_-_79536166 | 2.23 |
ENSMUST00000029386.14
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr2_-_165996716 | 2.22 |
ENSMUST00000139266.2
|
Sulf2
|
sulfatase 2 |
| chr1_-_24139263 | 2.20 |
ENSMUST00000187369.7
ENSMUST00000187752.7 ENSMUST00000186999.7 |
Fam135a
|
family with sequence similarity 135, member A |
| chr5_-_76452365 | 2.19 |
ENSMUST00000075159.5
|
Clock
|
circadian locomotor output cycles kaput |
| chr2_+_118998235 | 2.19 |
ENSMUST00000057454.4
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr2_-_26012751 | 2.18 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr10_-_94780695 | 2.18 |
ENSMUST00000099337.5
|
Plxnc1
|
plexin C1 |
| chr14_-_34224620 | 2.17 |
ENSMUST00000049005.15
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr6_-_119521243 | 2.17 |
ENSMUST00000119369.2
ENSMUST00000178696.8 |
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr4_+_40722461 | 2.17 |
ENSMUST00000030118.10
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr2_+_24944457 | 2.16 |
ENSMUST00000140737.8
|
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr13_+_47347301 | 2.14 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr8_-_123885007 | 2.13 |
ENSMUST00000000755.15
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr13_-_58363431 | 2.13 |
ENSMUST00000076454.8
ENSMUST00000058735.12 |
Ubqln1
|
ubiquilin 1 |
| chr15_-_76079891 | 2.11 |
ENSMUST00000023226.13
|
Plec
|
plectin |
| chr11_-_20781009 | 2.11 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr2_-_122544535 | 2.11 |
ENSMUST00000005952.11
|
Slc30a4
|
solute carrier family 30 (zinc transporter), member 4 |
| chr19_+_53933271 | 2.10 |
ENSMUST00000025932.9
|
Shoc2
|
Shoc2, leucine rich repeat scaffold protein |
| chr5_+_118114826 | 2.09 |
ENSMUST00000035579.10
|
Fbxo21
|
F-box protein 21 |
| chr13_+_32985990 | 2.09 |
ENSMUST00000021832.7
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr14_+_122771734 | 2.08 |
ENSMUST00000154206.8
ENSMUST00000038374.13 ENSMUST00000135578.8 |
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide |
| chr17_+_35780977 | 2.06 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr14_-_45767421 | 2.05 |
ENSMUST00000150660.3
|
Fermt2
|
fermitin family member 2 |
| chr6_-_134543876 | 2.05 |
ENSMUST00000032322.15
ENSMUST00000126836.4 |
Lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr13_-_96028412 | 2.04 |
ENSMUST00000068603.8
|
Iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr8_-_25528972 | 2.04 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr6_+_54572096 | 2.03 |
ENSMUST00000119706.8
|
Plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr1_+_131890679 | 2.02 |
ENSMUST00000191034.2
ENSMUST00000177943.8 |
Gm29103
Slc45a3
|
predicted gene 29103 solute carrier family 45, member 3 |
| chr12_-_75782406 | 2.02 |
ENSMUST00000220285.2
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr11_+_53241561 | 2.01 |
ENSMUST00000060945.12
|
Aff4
|
AF4/FMR2 family, member 4 |
| chr1_+_63485332 | 2.00 |
ENSMUST00000114103.8
ENSMUST00000114107.3 |
Adam23
|
a disintegrin and metallopeptidase domain 23 |
| chr4_-_149391963 | 2.00 |
ENSMUST00000055647.15
ENSMUST00000030806.6 ENSMUST00000238956.2 ENSMUST00000060537.13 |
Kif1b
|
kinesin family member 1B |
| chr1_-_52766615 | 2.00 |
ENSMUST00000156876.8
ENSMUST00000087701.4 |
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr16_-_11727262 | 1.99 |
ENSMUST00000127972.8
ENSMUST00000121750.2 ENSMUST00000096272.11 ENSMUST00000073371.7 |
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr5_+_118114795 | 1.99 |
ENSMUST00000202447.4
|
Fbxo21
|
F-box protein 21 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 11.9 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 2.6 | 10.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 2.5 | 7.6 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 2.3 | 6.8 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 2.2 | 6.5 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 2.2 | 13.0 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 2.1 | 12.9 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 1.7 | 7.0 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 1.4 | 10.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.4 | 5.6 | GO:1904412 | regulation of cardiac ventricle development(GO:1904412) |
| 1.2 | 3.7 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 1.2 | 6.0 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) |
| 1.2 | 7.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 1.2 | 10.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.2 | 8.1 | GO:0003383 | apical constriction(GO:0003383) |
| 1.1 | 5.7 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.1 | 4.4 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.1 | 4.2 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 1.0 | 3.0 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 1.0 | 3.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.0 | 3.0 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 1.0 | 3.0 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 1.0 | 2.9 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.9 | 3.8 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.9 | 2.8 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.9 | 2.8 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.9 | 6.4 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.9 | 2.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.9 | 2.6 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.9 | 3.5 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.9 | 2.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.8 | 5.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.8 | 5.0 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.8 | 8.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.8 | 4.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.8 | 2.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.8 | 2.4 | GO:0060365 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 0.8 | 7.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.8 | 2.4 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.8 | 2.4 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.8 | 2.4 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.8 | 3.9 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.8 | 5.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.8 | 3.8 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.7 | 5.2 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.7 | 5.9 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.7 | 3.7 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.7 | 2.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.7 | 2.2 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.7 | 1.4 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.7 | 2.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.7 | 4.9 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.7 | 1.3 | GO:0045818 | negative regulation of glycogen catabolic process(GO:0045818) |
| 0.7 | 4.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.7 | 2.0 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.6 | 4.5 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.6 | 7.7 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.6 | 1.9 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.6 | 2.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.6 | 5.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.6 | 2.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.6 | 1.9 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.6 | 1.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.6 | 2.4 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.6 | 1.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.6 | 3.0 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.6 | 1.8 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.6 | 1.8 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.6 | 2.3 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.6 | 2.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.6 | 2.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.5 | 11.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.5 | 15.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 5.9 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.5 | 2.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.5 | 1.6 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.5 | 1.6 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.5 | 0.5 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.5 | 6.2 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.5 | 1.5 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.5 | 5.5 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.5 | 1.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.5 | 2.5 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.5 | 1.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.5 | 2.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.5 | 2.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.5 | 3.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.5 | 1.4 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.5 | 0.9 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.5 | 3.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 2.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.4 | 1.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 1.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.4 | 1.8 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.4 | 1.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.4 | 3.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.4 | 3.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 1.7 | GO:0046959 | habituation(GO:0046959) |
| 0.4 | 1.2 | GO:0015881 | creatine transport(GO:0015881) |
| 0.4 | 1.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.4 | 2.4 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.4 | 1.6 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.4 | 0.4 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.4 | 1.2 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.4 | 0.8 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.4 | 1.9 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.4 | 4.7 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.4 | 2.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 1.5 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.4 | 1.5 | GO:1902045 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.4 | 4.2 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 2.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.4 | 2.7 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.4 | 1.9 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.4 | 1.9 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.4 | 2.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.4 | 4.9 | GO:0007567 | parturition(GO:0007567) |
| 0.4 | 1.1 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.4 | 1.4 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.4 | 1.4 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.3 | 1.0 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.3 | 1.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.3 | 1.0 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 1.3 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.3 | 5.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 1.5 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 0.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.3 | 0.9 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.3 | 2.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.3 | 1.5 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.3 | 2.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.3 | 0.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 1.4 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.3 | 1.7 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.3 | 19.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.3 | 5.3 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.3 | 3.6 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.3 | 0.8 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 2.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 1.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.3 | 5.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.3 | 1.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 0.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 3.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 5.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 5.9 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.3 | 2.4 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.3 | 4.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.3 | 0.8 | GO:2000156 | regulation of constitutive secretory pathway(GO:1903433) regulation of retrograde vesicle-mediated transport, Golgi to ER(GO:2000156) |
| 0.3 | 1.0 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.3 | 2.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.3 | 0.8 | GO:0061646 | positive regulation of glutamate neurotransmitter secretion in response to membrane depolarization(GO:0061646) |
| 0.3 | 1.0 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 | 2.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 2.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.5 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.2 | 4.0 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 0.2 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 1.5 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 3.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 4.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 1.0 | GO:2000864 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.2 | 0.7 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 0.7 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.2 | 0.9 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 1.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 1.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 5.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 1.3 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 | 1.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 | 2.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 3.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 2.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 1.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 0.6 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 0.8 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 0.6 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 0.6 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 0.8 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 3.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.2 | 4.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 2.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 6.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 1.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 3.6 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 3.8 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 1.9 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.2 | 3.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 2.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 0.7 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.2 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 0.9 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 0.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 3.3 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 0.5 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.2 | 0.9 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.2 | 1.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.6 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 1.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 3.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 2.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 4.9 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.2 | 0.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 2.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 0.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.5 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.2 | 2.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.8 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 1.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.2 | 1.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 1.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 2.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 2.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 0.5 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 1.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.9 | GO:0061744 | adenosine to inosine editing(GO:0006382) motor behavior(GO:0061744) |
| 0.2 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 1.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 4.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.4 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.6 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.4 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 1.6 | GO:1902170 | cellular response to reactive nitrogen species(GO:1902170) |
| 0.1 | 1.0 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.5 | GO:0042335 | cuticle development(GO:0042335) cornification(GO:0070268) |
| 0.1 | 0.9 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.4 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.1 | 1.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.3 | GO:2000860 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 4.8 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.1 | 0.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.6 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.8 | GO:0090032 | negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.1 | 0.9 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 0.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 1.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 2.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.8 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 1.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.9 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 1.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 0.6 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.5 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 1.6 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.0 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 4.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.8 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 1.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.4 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.5 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.7 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.1 | 0.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 2.7 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.4 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 0.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 2.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 0.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 1.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 1.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 2.9 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 0.8 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 1.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 10.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 1.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 3.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.4 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.4 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 0.3 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 4.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 3.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 1.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.5 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.4 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 3.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 0.5 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 1.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.1 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 0.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 5.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.2 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 1.8 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.2 | GO:0036257 | multivesicular body organization(GO:0036257) multivesicular body assembly(GO:0036258) |
| 0.1 | 0.1 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 0.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.6 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 1.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 2.2 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 4.5 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 0.3 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.1 | 3.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 0.2 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 2.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.2 | GO:0007442 | hindgut morphogenesis(GO:0007442) |
| 0.1 | 0.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 2.2 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.9 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.2 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 2.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.1 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 0.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.3 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.7 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 1.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.5 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.4 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 3.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.6 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.2 | GO:1904798 | negative regulation of lymphangiogenesis(GO:1901491) regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 0.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.2 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.1 | 1.1 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.1 | 0.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.1 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 1.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.3 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 5.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 2.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.0 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.8 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.5 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.2 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.1 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.6 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 1.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.9 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.6 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0060540 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 2.4 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.6 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 1.0 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 1.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.2 | GO:2000395 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 3.0 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 1.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.2 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 1.5 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.6 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.4 | GO:0051196 | regulation of coenzyme metabolic process(GO:0051196) |
| 0.0 | 0.3 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 1.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 3.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 1.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.5 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.8 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.4 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 2.5 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.0 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:1903977 | positive regulation of Schwann cell migration(GO:1900149) positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.0 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.2 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.7 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.3 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 11.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.0 | 3.0 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 1.0 | 4.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.9 | 3.7 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.8 | 7.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.8 | 2.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.7 | 14.1 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.7 | 2.0 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.6 | 4.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.5 | 2.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.4 | 1.8 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 0.8 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.4 | 2.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.4 | 0.8 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 0.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.4 | 7.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.4 | 11.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 2.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.4 | 1.5 | GO:0071149 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 9.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 3.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 1.7 | GO:0044308 | axonal spine(GO:0044308) |
| 0.3 | 1.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.3 | 5.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 9.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.3 | 3.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 13.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 0.8 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.3 | 1.9 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.3 | 1.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.3 | 4.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.3 | 2.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.2 | 0.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.9 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 2.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 6.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 1.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 18.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 4.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 2.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 0.5 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 4.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.2 | 1.5 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.9 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 8.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 5.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 1.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 2.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 7.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.6 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.2 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 2.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 2.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 5.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 11.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.2 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.1 | 3.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 4.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 2.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 2.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.4 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 0.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.7 | GO:0030897 | HOPS complex(GO:0030897) FHF complex(GO:0070695) |
| 0.1 | 2.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.7 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 5.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 3.6 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 3.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.9 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 7.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.8 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 2.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 10.7 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 1.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 6.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 2.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 2.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 5.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 22.4 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 8.5 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 3.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 39.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 2.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 2.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 4.3 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 8.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 7.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 2.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 2.2 | 6.5 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitric oxide binding(GO:0070026) |
| 2.1 | 8.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 2.0 | 5.9 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 1.8 | 1.8 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 1.6 | 11.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.4 | 5.4 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 1.4 | 6.8 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 1.3 | 4.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.3 | 11.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.2 | 3.7 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 1.2 | 4.8 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.2 | 4.7 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.2 | 8.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 1.1 | 3.4 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 1.1 | 15.4 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 1.1 | 4.4 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.1 | 6.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.1 | 4.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 1.1 | 3.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 1.0 | 4.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 1.0 | 5.2 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 1.0 | 6.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.0 | 5.7 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.0 | 4.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.0 | 3.8 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.9 | 2.8 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.9 | 2.8 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.9 | 4.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.9 | 2.6 | GO:0005369 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.9 | 2.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.8 | 2.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.8 | 1.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.8 | 7.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.8 | 2.4 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.8 | 4.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.8 | 2.4 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.7 | 3.7 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.7 | 13.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.7 | 2.2 | GO:0047945 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.7 | 5.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.7 | 5.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 2.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.7 | 2.0 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.6 | 2.6 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.6 | 0.6 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.6 | 3.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 3.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.6 | 3.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.6 | 2.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.6 | 3.0 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.6 | 2.4 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.6 | 4.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.6 | 5.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.6 | 3.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.6 | 2.2 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.5 | 3.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 1.5 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.5 | 5.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 2.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.5 | 2.0 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.5 | 2.4 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.5 | 1.9 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 2.8 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.5 | 2.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.5 | 3.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.4 | 1.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.4 | 3.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.4 | 4.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 4.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.4 | 10.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 5.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 2.5 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.4 | 1.2 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.4 | 1.6 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.4 | 2.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 4.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 2.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.4 | 1.9 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 2.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 1.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.4 | 4.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 2.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.3 | 1.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 1.6 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.3 | 4.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 2.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 4.0 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.3 | 1.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 0.9 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 4.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 9.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.3 | 2.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.3 | 1.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 0.9 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.3 | 8.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.3 | 0.9 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.3 | 2.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 2.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 1.7 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 1.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.3 | 4.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 2.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 2.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 2.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 1.0 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 0.9 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 5.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 0.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 3.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 12.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 1.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 1.3 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.2 | 3.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 8.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 5.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 4.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 3.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.2 | 4.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 1.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 5.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.7 | GO:0004080 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.2 | 1.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 1.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 0.4 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 4.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 2.9 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.2 | 2.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 2.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 1.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 2.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 1.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.2 | 2.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 1.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 3.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 8.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 2.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 6.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.9 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 0.9 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.2 | 0.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 2.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 4.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 6.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 1.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 7.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 2.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 6.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.3 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.5 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.3 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 1.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 11.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 7.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 2.9 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 0.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 2.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.3 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 1.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 5.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 5.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 1.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 7.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 1.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 2.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 2.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 5.2 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.6 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.6 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 2.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 2.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 1.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.0 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 7.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 2.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.9 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 1.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 4.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 2.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 3.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 2.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.3 | 13.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 5.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 6.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 4.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 4.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 15.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 8.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 5.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 3.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 4.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 5.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 4.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 21.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.9 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 11.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 3.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 6.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 19.9 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.5 | 12.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 4.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 9.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 6.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 4.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 5.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 9.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 5.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 3.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 4.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 11.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 5.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 3.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 9.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 2.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 8.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.7 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.2 | 18.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 7.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 4.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 9.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 8.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 5.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 5.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 5.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.2 | 2.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 7.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 4.3 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.2 | 3.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 1.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 4.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 7.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 3.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 1.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 7.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 3.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 4.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 4.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 6.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 4.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 6.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 7.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 5.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 1.4 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 2.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 4.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 5.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.6 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |