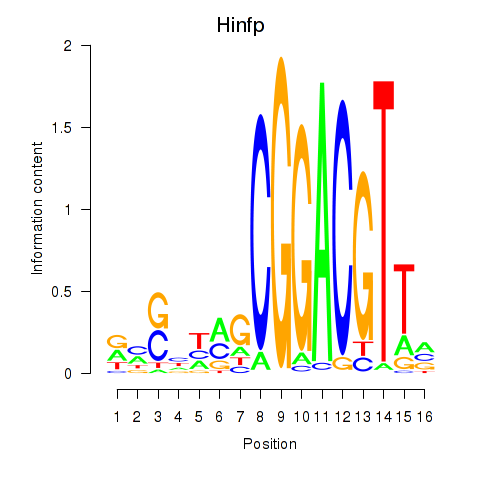
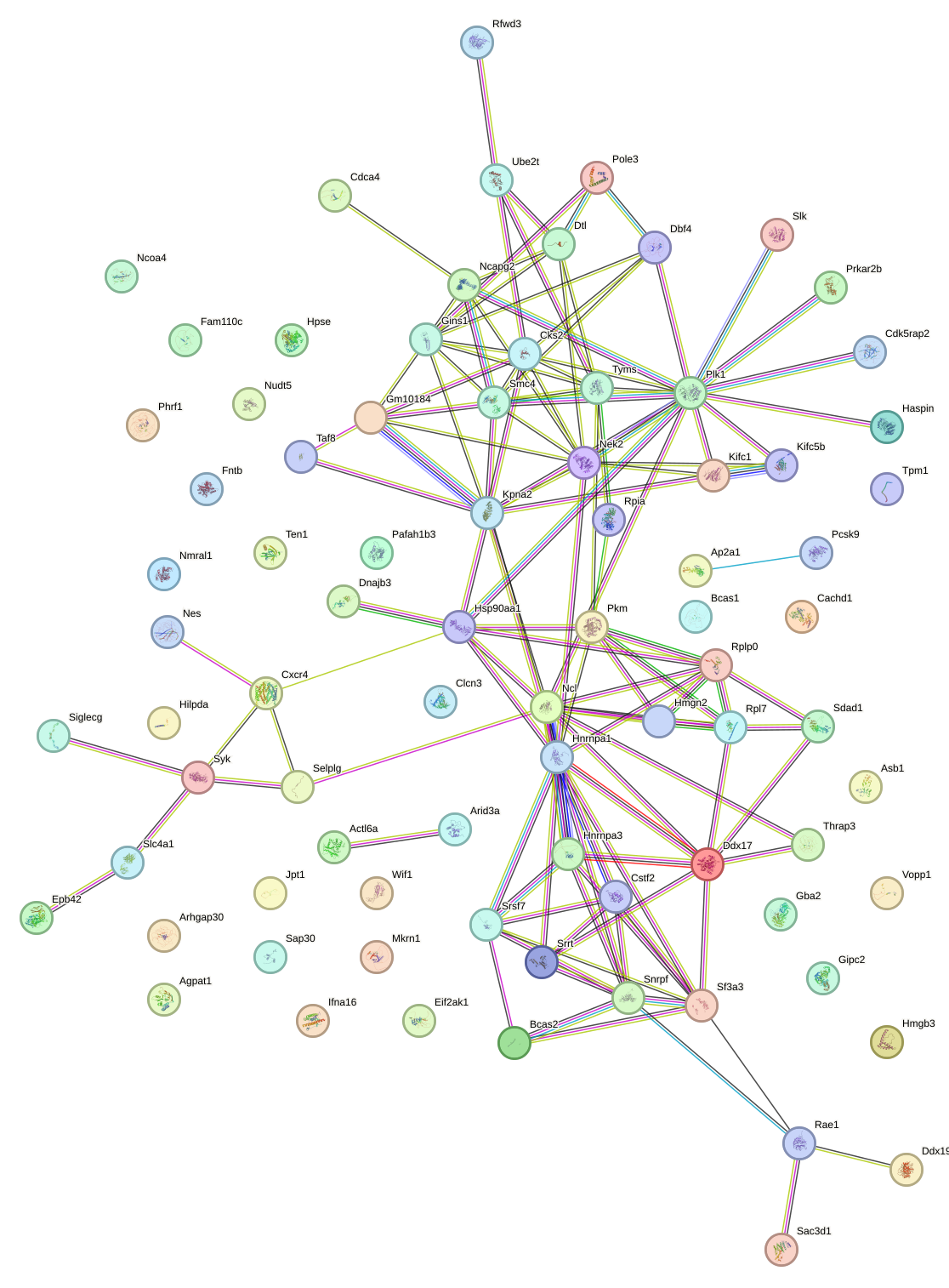
|
chr11_-_102255999
Show fit
|
22.53 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1
|
|
chr12_-_32111214
Show fit
|
11.57 |
ENSMUST00000003079.12
ENSMUST00000036497.16
|
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta
|
|
chr2_-_120867529
Show fit
|
11.32 |
ENSMUST00000102490.10
|
Epb42
|
erythrocyte membrane protein band 4.2
|
|
chr15_+_103148824
Show fit
|
10.41 |
ENSMUST00000036004.16
ENSMUST00000087351.9
ENSMUST00000231141.2
|
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1
|
|
chr2_-_120867232
Show fit
|
10.11 |
ENSMUST00000023987.6
|
Epb42
|
erythrocyte membrane protein band 4.2
|
|
chr7_+_121758646
Show fit
|
8.93 |
ENSMUST00000033154.8
ENSMUST00000205901.2
|
Plk1
|
polo like kinase 1
|
|
chr1_-_128520002
Show fit
|
8.93 |
ENSMUST00000052172.7
ENSMUST00000142893.2
|
Cxcr4
|
chemokine (C-X-C motif) receptor 4
|
|
chr3_-_151871867
Show fit
|
8.66 |
ENSMUST00000046614.10
|
Gipc2
|
GIPC PDZ domain containing family, member 2
|
|
chr2_+_84670543
Show fit
|
8.58 |
ENSMUST00000111624.8
|
Slc43a1
|
solute carrier family 43, member 1
|
|
chr6_-_39396691
Show fit
|
8.21 |
ENSMUST00000146785.8
ENSMUST00000114823.8
|
Mkrn1
|
makorin, ring finger protein, 1
|
|
chr2_+_84670956
Show fit
|
8.15 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1
|
|
chr1_-_88133472
Show fit
|
7.93 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3
|
|
chr5_+_33815910
Show fit
|
7.15 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr11_+_116089678
Show fit
|
7.05 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit
|
|
chr2_-_156848923
Show fit
|
6.88 |
ENSMUST00000146413.8
ENSMUST00000103129.9
ENSMUST00000103130.8
|
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component
|
|
chr6_-_57802131
Show fit
|
6.37 |
ENSMUST00000204878.3
ENSMUST00000145608.7
ENSMUST00000203212.3
ENSMUST00000114297.5
|
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1
|
|
chr12_+_111383864
Show fit
|
6.26 |
ENSMUST00000220537.2
ENSMUST00000223050.2
ENSMUST00000072646.8
ENSMUST00000223431.2
ENSMUST00000221144.2
ENSMUST00000222437.2
|
Exoc3l4
|
exocyst complex component 3-like 4
|
|
chr10_-_93425553
Show fit
|
6.25 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr5_+_115697526
Show fit
|
6.15 |
ENSMUST00000086519.12
ENSMUST00000156359.2
ENSMUST00000152976.2
|
Rplp0
|
ribosomal protein, large, P0
|
|
chrX_+_70599524
Show fit
|
6.14 |
ENSMUST00000072699.13
ENSMUST00000114582.9
ENSMUST00000015361.11
ENSMUST00000088874.10
|
Hmgb3
|
high mobility group box 3
|
|
chr9_+_59564482
Show fit
|
6.01 |
ENSMUST00000216620.2
ENSMUST00000217038.2
|
Pkm
|
pyruvate kinase, muscle
|
|
chr6_+_29272625
Show fit
|
6.01 |
ENSMUST00000054445.9
|
Hilpda
|
hypoxia inducible lipid droplet associated
|
|
chr5_-_8472582
Show fit
|
5.89 |
ENSMUST00000168500.8
ENSMUST00000002368.16
|
Dbf4
|
DBF4 zinc finger
|
|
chr8_-_71219299
Show fit
|
5.51 |
ENSMUST00000222087.4
|
Ifi30
|
interferon gamma inducible protein 30
|
|
chr17_+_27136065
Show fit
|
5.36 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B
|
|
chr12_+_76884182
Show fit
|
5.34 |
ENSMUST00000041008.10
|
Fntb
|
farnesyltransferase, CAAX box, beta
|
|
chr1_+_191553556
Show fit
|
5.26 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2
|
|
chr12_-_112792971
Show fit
|
5.13 |
ENSMUST00000062092.7
ENSMUST00000220899.2
|
Cdca4
|
cell division cycle associated 4
|
|
chr13_+_51799268
Show fit
|
5.04 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr17_-_80514725
Show fit
|
5.00 |
ENSMUST00000234696.2
ENSMUST00000235069.2
ENSMUST00000063417.11
|
Srsf7
|
serine and arginine-rich splicing factor 7
|
|
chr6_-_70769135
Show fit
|
4.92 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A
|
|
chr5_-_8472696
Show fit
|
4.83 |
ENSMUST00000171808.8
|
Dbf4
|
DBF4 zinc finger
|
|
chr5_-_30278552
Show fit
|
4.83 |
ENSMUST00000198095.2
ENSMUST00000196872.2
ENSMUST00000026846.11
|
Tyms
|
thymidylate synthase
|
|
chr8_-_57940834
Show fit
|
4.82 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide
|
|
chr6_+_29272463
Show fit
|
4.75 |
ENSMUST00000115289.2
|
Hilpda
|
hypoxia inducible lipid droplet associated
|
|
chr1_+_134890288
Show fit
|
4.58 |
ENSMUST00000027687.8
|
Ube2t
|
ubiquitin-conjugating enzyme E2T
|
|
chr11_-_73029070
Show fit
|
4.50 |
ENSMUST00000052140.3
|
Haspin
|
histone H3 associated protein kinase
|
|
chr4_-_133695264
Show fit
|
4.47 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2
|
|
chr7_-_24997291
Show fit
|
4.42 |
ENSMUST00000148150.8
ENSMUST00000155118.2
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3
|
|
chr19_+_47568041
Show fit
|
4.29 |
ENSMUST00000026043.12
|
Slk
|
STE20-like kinase
|
|
chr11_-_115405200
Show fit
|
4.24 |
ENSMUST00000021083.7
|
Jpt1
|
Jupiter microtubule associated homolog 1
|
|
chr4_-_106321363
Show fit
|
4.08 |
ENSMUST00000049507.6
|
Pcsk9
|
proprotein convertase subtilisin/kexin type 9
|
|
chr12_+_31123860
Show fit
|
4.07 |
ENSMUST00000041133.10
|
Fam110c
|
family with sequence similarity 110, member C
|
|
chr7_-_44578834
Show fit
|
4.06 |
ENSMUST00000107857.11
ENSMUST00000167930.8
ENSMUST00000085399.13
ENSMUST00000166972.9
|
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit
|
|
chr13_+_52737118
Show fit
|
4.04 |
ENSMUST00000120135.8
|
Syk
|
spleen tyrosine kinase
|
|
chrX_+_163202778
Show fit
|
3.98 |
ENSMUST00000208741.2
ENSMUST00000033754.15
ENSMUST00000208697.2
ENSMUST00000208261.2
|
Piga
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr8_-_111724409
Show fit
|
3.90 |
ENSMUST00000040416.8
|
Ddx19a
|
DEAD box helicase 19a
|
|
chr3_+_68912043
Show fit
|
3.86 |
ENSMUST00000042901.15
|
Smc4
|
structural maintenance of chromosomes 4
|
|
chr9_-_66956425
Show fit
|
3.84 |
ENSMUST00000113687.8
ENSMUST00000113693.8
ENSMUST00000113701.8
ENSMUST00000034928.12
ENSMUST00000113685.10
ENSMUST00000030185.5
ENSMUST00000050905.16
ENSMUST00000113705.8
ENSMUST00000113697.8
ENSMUST00000113707.9
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr5_-_92457862
Show fit
|
3.81 |
ENSMUST00000031364.5
|
Sdad1
|
SDA1 domain containing 1
|
|
chr19_+_53131187
Show fit
|
3.81 |
ENSMUST00000050096.15
ENSMUST00000237832.2
|
Add3
|
adducin 3 (gamma)
|
|
chr14_+_31881822
Show fit
|
3.75 |
ENSMUST00000163336.8
ENSMUST00000169722.8
ENSMUST00000168385.8
|
Ncoa4
|
nuclear receptor coactivator 4
|
|
chr2_-_170269748
Show fit
|
3.74 |
ENSMUST00000013667.3
ENSMUST00000109152.9
ENSMUST00000068137.11
|
Bcas1
|
breast carcinoma amplified sequence 1
|
|
chr5_+_143803540
Show fit
|
3.59 |
ENSMUST00000100487.6
|
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1
|
|
chr1_+_171216480
Show fit
|
3.53 |
ENSMUST00000056449.9
|
Arhgap30
|
Rho GTPase activating protein 30
|
|
chr2_+_172841907
Show fit
|
3.49 |
ENSMUST00000029013.10
ENSMUST00000132212.2
|
Rae1
|
ribonucleic acid export 1
|
|
chr5_+_33815892
Show fit
|
3.44 |
ENSMUST00000152847.8
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr11_+_69737437
Show fit
|
3.37 |
ENSMUST00000152566.8
ENSMUST00000108633.9
|
Plscr3
|
phospholipid scramblase 3
|
|
chr12_+_116369017
Show fit
|
3.36 |
ENSMUST00000084828.5
ENSMUST00000222469.2
ENSMUST00000221114.2
ENSMUST00000221970.2
|
Ncapg2
|
non-SMC condensin II complex, subunit G2
|
|
chr10_+_120869865
Show fit
|
3.35 |
ENSMUST00000020439.11
ENSMUST00000175867.2
|
Wif1
|
Wnt inhibitory factor 1
|
|
chr4_-_126096376
Show fit
|
3.20 |
ENSMUST00000106142.8
ENSMUST00000169403.8
ENSMUST00000130334.2
|
Thrap3
|
thyroid hormone receptor associated protein 3
|
|
chr3_+_87878378
Show fit
|
3.17 |
ENSMUST00000090973.12
|
Nes
|
nestin
|
|
chr4_+_156194427
Show fit
|
3.13 |
ENSMUST00000072554.13
ENSMUST00000169550.8
ENSMUST00000105576.2
|
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene
|
|
chr5_-_100867520
Show fit
|
3.11 |
ENSMUST00000112908.2
ENSMUST00000045617.15
|
Hpse
|
heparanase
|
|
chr9_-_66951114
Show fit
|
3.11 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr10_+_79763164
Show fit
|
3.10 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr16_-_4536992
Show fit
|
3.07 |
ENSMUST00000115851.10
|
Nmral1
|
NmrA-like family domain containing 1
|
|
chr3_+_87878436
Show fit
|
3.06 |
ENSMUST00000160694.2
|
Nes
|
nestin
|
|
chr7_+_43057611
Show fit
|
3.05 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G
|
|
chr2_+_5849828
Show fit
|
3.01 |
ENSMUST00000026927.10
ENSMUST00000179748.8
|
Nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5
|
|
chr1_-_191307648
Show fit
|
3.01 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase
|
|
chr5_-_137305895
Show fit
|
2.97 |
ENSMUST00000199243.5
ENSMUST00000197466.5
ENSMUST00000040873.12
|
Srrt
|
serrate RNA effector molecule homolog (Arabidopsis)
|
|
chr8_+_84442133
Show fit
|
2.93 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a
|
|
chr1_-_86287080
Show fit
|
2.92 |
ENSMUST00000027438.8
|
Ncl
|
nucleolin
|
|
chr7_+_46495521
Show fit
|
2.91 |
ENSMUST00000133062.2
|
Ldha
|
lactate dehydrogenase A
|
|
chr2_+_5850053
Show fit
|
2.88 |
ENSMUST00000127116.7
ENSMUST00000194933.2
|
Nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5
|
|
chr8_-_112026854
Show fit
|
2.87 |
ENSMUST00000038739.5
|
Rfwd3
|
ring finger and WD repeat domain 3
|
|
chr11_-_106890307
Show fit
|
2.84 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2
|
|
chr4_-_43578823
Show fit
|
2.84 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2
|
|
chr1_+_91468796
Show fit
|
2.82 |
ENSMUST00000188081.7
ENSMUST00000188879.2
|
Asb1
|
ankyrin repeat and SOCS box-containing 1
|
|
chrX_+_135567124
Show fit
|
2.80 |
ENSMUST00000060904.11
ENSMUST00000113100.2
ENSMUST00000128040.2
|
Tceal3
|
transcription elongation factor A (SII)-like 3
|
|
chr12_-_110662765
Show fit
|
2.78 |
ENSMUST00000094361.11
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1
|
|
chr4_+_152123772
Show fit
|
2.73 |
ENSMUST00000084116.13
ENSMUST00000103197.5
|
Nol9
|
nucleolar protein 9
|
|
chr4_-_70328659
Show fit
|
2.69 |
ENSMUST00000144099.8
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2
|
|
chrX_+_132959905
Show fit
|
2.68 |
ENSMUST00000113287.8
ENSMUST00000033609.9
ENSMUST00000113286.8
|
Cstf2
|
cleavage stimulation factor, 3' pre-RNA subunit 2
|
|
chr11_-_106890195
Show fit
|
2.63 |
ENSMUST00000106768.2
ENSMUST00000144834.8
|
Kpna2
|
karyopherin (importin) alpha 2
|
|
chr5_-_113957362
Show fit
|
2.61 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand
|
|
chr1_-_16174387
Show fit
|
2.57 |
ENSMUST00000149566.2
|
Rpl7
|
ribosomal protein L7
|
|
chr4_+_100633860
Show fit
|
2.51 |
ENSMUST00000030257.15
ENSMUST00000097955.3
|
Cachd1
|
cache domain containing 1
|
|
chr2_+_75489596
Show fit
|
2.50 |
ENSMUST00000111964.8
ENSMUST00000111962.8
ENSMUST00000111961.8
ENSMUST00000164947.9
ENSMUST00000090792.11
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3
|
|
chr8_+_84441854
Show fit
|
2.46 |
ENSMUST00000172396.8
|
Ddx39a
|
DEAD box helicase 39a
|
|
chr3_+_32762656
Show fit
|
2.43 |
ENSMUST00000029214.14
|
Actl6a
|
actin-like 6A
|
|
chr5_-_113957318
Show fit
|
2.43 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand
|
|
chr7_+_140808680
Show fit
|
2.40 |
ENSMUST00000106027.9
|
Phrf1
|
PHD and ring finger domains 1
|
|
chr19_-_6168518
Show fit
|
2.39 |
ENSMUST00000113533.3
|
Sac3d1
|
SAC3 domain containing 1
|
|
chr17_-_47813201
Show fit
|
2.38 |
ENSMUST00000233174.2
ENSMUST00000233121.2
ENSMUST00000067103.4
|
Taf8
|
TATA-box binding protein associated factor 8
|
|
chr4_-_62443273
Show fit
|
2.38 |
ENSMUST00000030091.10
|
Pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit)
|
|
chr9_-_66951151
Show fit
|
2.37 |
ENSMUST00000113696.8
|
Tpm1
|
tropomyosin 1, alpha
|
|
chrX_+_163202852
Show fit
|
2.37 |
ENSMUST00000112255.8
|
Piga
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr17_+_34823236
Show fit
|
2.36 |
ENSMUST00000174041.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr4_-_126096551
Show fit
|
2.34 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3
|
|
chr4_+_124608569
Show fit
|
2.33 |
ENSMUST00000030734.5
|
Sf3a3
|
splicing factor 3a, subunit 3
|
|
chr8_-_61436249
Show fit
|
2.28 |
ENSMUST00000004430.14
ENSMUST00000110301.2
ENSMUST00000093490.9
|
Clcn3
|
chloride channel, voltage-sensitive 3
|
|
chr8_+_84441806
Show fit
|
2.28 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a
|
|
chr15_-_79430742
Show fit
|
2.25 |
ENSMUST00000231053.2
ENSMUST00000229431.2
|
Ddx17
|
DEAD box helicase 17
|
|
chr4_-_88595161
Show fit
|
2.24 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16
|
|
chr7_+_46495256
Show fit
|
2.24 |
ENSMUST00000048209.16
ENSMUST00000210815.2
ENSMUST00000125862.8
ENSMUST00000210968.2
ENSMUST00000092621.12
ENSMUST00000210467.2
|
Ldha
|
lactate dehydrogenase A
|
|
chr3_+_103078971
Show fit
|
2.22 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2
|
|
chr2_+_150751475
Show fit
|
2.20 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog)
|
|
chr4_-_116982804
Show fit
|
2.18 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19
|
|
chr18_+_36877709
Show fit
|
2.15 |
ENSMUST00000007042.6
ENSMUST00000237095.2
|
Ik
|
IK cytokine
|
|
chr5_-_92457753
Show fit
|
2.15 |
ENSMUST00000201143.2
|
Sdad1
|
SDA1 domain containing 1
|
|
chr5_+_129578285
Show fit
|
2.15 |
ENSMUST00000053737.9
|
Sfswap
|
splicing factor SWAP
|
|
chr4_+_43578707
Show fit
|
2.14 |
ENSMUST00000107886.9
ENSMUST00000117140.8
|
Rgp1
|
RAB6A GEF compex partner 1
|
|
chr19_-_34143437
Show fit
|
2.14 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22
|
|
chr13_-_95661726
Show fit
|
2.13 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr4_-_43578636
Show fit
|
2.13 |
ENSMUST00000130443.2
|
Gba2
|
glucosidase beta 2
|
|
chr6_-_113717689
Show fit
|
2.12 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component
|
|
chr3_+_32763313
Show fit
|
2.03 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A
|
|
chr14_+_30723340
Show fit
|
2.02 |
ENSMUST00000168584.9
ENSMUST00000226378.2
|
Glt8d1
|
glycosyltransferase 8 domain containing 1
|
|
chr11_+_97917520
Show fit
|
2.02 |
ENSMUST00000092425.11
|
Rpl19
|
ribosomal protein L19
|
|
chr9_-_66951234
Show fit
|
2.02 |
ENSMUST00000113690.8
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr1_-_97589675
Show fit
|
2.01 |
ENSMUST00000053033.14
ENSMUST00000149927.2
|
Macir
|
macrophage immunometabolism regulator
|
|
chr7_-_92523396
Show fit
|
2.01 |
ENSMUST00000209074.2
ENSMUST00000208356.2
ENSMUST00000032877.11
|
Ddias
|
DNA damage-induced apoptosis suppressor
|
|
chr9_-_66950991
Show fit
|
2.00 |
ENSMUST00000113689.8
ENSMUST00000113684.8
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr19_+_53588808
Show fit
|
2.00 |
ENSMUST00000025930.10
|
Smc3
|
structural maintenance of chromosomes 3
|
|
chr6_+_47930324
Show fit
|
1.98 |
ENSMUST00000101445.11
|
Zfp956
|
zinc finger protein 956
|
|
chrX_-_74460168
Show fit
|
1.96 |
ENSMUST00000033543.14
ENSMUST00000149863.3
ENSMUST00000114081.2
|
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4
mature T cell proliferation 1
|
|
chr11_-_117859997
Show fit
|
1.94 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3
|
|
chr17_+_24633614
Show fit
|
1.93 |
ENSMUST00000115371.9
ENSMUST00000088512.13
ENSMUST00000163717.2
|
Rnps1
|
RNA binding protein with serine rich domain 1
|
|
chr3_+_151916106
Show fit
|
1.93 |
ENSMUST00000199202.5
ENSMUST00000200524.5
ENSMUST00000198227.5
ENSMUST00000196739.5
ENSMUST00000196695.5
ENSMUST00000106121.6
|
Fubp1
|
far upstream element (FUSE) binding protein 1
|
|
chr11_-_79145489
Show fit
|
1.91 |
ENSMUST00000017821.12
|
Wsb1
|
WD repeat and SOCS box-containing 1
|
|
chr1_+_139382485
Show fit
|
1.91 |
ENSMUST00000200083.5
ENSMUST00000053364.12
|
Aspm
|
abnormal spindle microtubule assembly
|
|
chr12_+_84034628
Show fit
|
1.90 |
ENSMUST00000021649.8
|
Acot2
|
acyl-CoA thioesterase 2
|
|
chr15_+_84807582
Show fit
|
1.84 |
ENSMUST00000165443.4
|
Nup50
|
nucleoporin 50
|
|
chr8_+_75742850
Show fit
|
1.82 |
ENSMUST00000109940.2
|
Hmgxb4
|
HMG box domain containing 4
|
|
chr9_+_99125420
Show fit
|
1.82 |
ENSMUST00000185799.7
ENSMUST00000093795.10
ENSMUST00000190715.7
ENSMUST00000191335.7
ENSMUST00000190078.7
|
Cep70
|
centrosomal protein 70
|
|
chr14_-_103336990
Show fit
|
1.77 |
ENSMUST00000022720.15
ENSMUST00000144141.8
|
Fbxl3
|
F-box and leucine-rich repeat protein 3
|
|
chr17_-_23892798
Show fit
|
1.76 |
ENSMUST00000047436.11
ENSMUST00000115490.9
ENSMUST00000095579.11
|
Thoc6
|
THO complex 6
|
|
chr8_-_103512274
Show fit
|
1.71 |
ENSMUST00000075190.5
|
Cdh11
|
cadherin 11
|
|
chr14_+_30723371
Show fit
|
1.71 |
ENSMUST00000022476.9
|
Glt8d1
|
glycosyltransferase 8 domain containing 1
|
|
chr12_+_51394812
Show fit
|
1.69 |
ENSMUST00000054308.13
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase
|
|
chr13_-_99027482
Show fit
|
1.68 |
ENSMUST00000179301.8
ENSMUST00000179271.2
|
Tnpo1
|
transportin 1
|
|
chr10_+_13428638
Show fit
|
1.68 |
ENSMUST00000019944.9
|
Adat2
|
adenosine deaminase, tRNA-specific 2
|
|
chr15_-_31601652
Show fit
|
1.67 |
ENSMUST00000161266.2
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon)
|
|
chr15_-_77854988
Show fit
|
1.66 |
ENSMUST00000100484.6
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr15_+_84807640
Show fit
|
1.66 |
ENSMUST00000230411.2
|
Nup50
|
nucleoporin 50
|
|
chr11_-_75060345
Show fit
|
1.65 |
ENSMUST00000055619.5
|
Hic1
|
hypermethylated in cancer 1
|
|
chr2_+_28423367
Show fit
|
1.65 |
ENSMUST00000113893.8
ENSMUST00000100241.10
|
Ralgds
|
ral guanine nucleotide dissociation stimulator
|
|
chrX_+_156482116
Show fit
|
1.64 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked
|
|
chr17_+_28059036
Show fit
|
1.64 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C
|
|
chr5_-_77457895
Show fit
|
1.63 |
ENSMUST00000047860.9
|
Noa1
|
nitric oxide associated 1
|
|
chr10_+_85707687
Show fit
|
1.61 |
ENSMUST00000001836.11
|
Pwp1
|
PWP1 homolog, endonuclein
|
|
chr5_-_113910571
Show fit
|
1.54 |
ENSMUST00000019118.8
|
Sart3
|
squamous cell carcinoma antigen recognized by T cells 3
|
|
chr2_+_31578537
Show fit
|
1.52 |
ENSMUST00000075759.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr12_+_80992276
Show fit
|
1.51 |
ENSMUST00000094693.11
|
Srsf5
|
serine and arginine-rich splicing factor 5
|
|
chr9_-_66951025
Show fit
|
1.48 |
ENSMUST00000113695.8
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr12_-_110662723
Show fit
|
1.48 |
ENSMUST00000021698.13
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1
|
|
chr11_-_98291278
Show fit
|
1.46 |
ENSMUST00000090827.12
|
Pgap3
|
post-GPI attachment to proteins 3
|
|
chr19_-_45738002
Show fit
|
1.45 |
ENSMUST00000070215.8
|
Npm3
|
nucleoplasmin 3
|
|
chr11_+_83189831
Show fit
|
1.45 |
ENSMUST00000176944.8
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr10_+_98942898
Show fit
|
1.43 |
ENSMUST00000020113.15
ENSMUST00000159228.8
ENSMUST00000159990.2
|
Poc1b
|
POC1 centriolar protein B
|
|
chr5_-_145103841
Show fit
|
1.43 |
ENSMUST00000031628.10
|
Ptcd1
|
pentatricopeptide repeat domain 1
|
|
chr17_+_6130205
Show fit
|
1.40 |
ENSMUST00000100955.3
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5
|
|
chr6_-_5496261
Show fit
|
1.39 |
ENSMUST00000203347.3
ENSMUST00000019721.7
|
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4
|
|
chr7_+_78545660
Show fit
|
1.37 |
ENSMUST00000107425.8
ENSMUST00000107421.8
|
Aen
|
apoptosis enhancing nuclease
|
|
chr5_+_30360246
Show fit
|
1.35 |
ENSMUST00000026841.15
ENSMUST00000123980.8
ENSMUST00000114783.6
ENSMUST00000114786.8
|
Hadhb
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta
|
|
chrX_+_156481906
Show fit
|
1.34 |
ENSMUST00000136141.2
ENSMUST00000190091.7
|
Smpx
|
small muscle protein, X-linked
|
|
chr12_+_70499869
Show fit
|
1.33 |
ENSMUST00000021471.13
|
Tmx1
|
thioredoxin-related transmembrane protein 1
|
|
chr19_-_36896999
Show fit
|
1.27 |
ENSMUST00000238948.2
ENSMUST00000057337.9
|
Fgfbp3
|
fibroblast growth factor binding protein 3
|
|
chr16_-_4782031
Show fit
|
1.24 |
ENSMUST00000023157.6
ENSMUST00000229765.2
|
Anks3
|
ankyrin repeat and sterile alpha motif domain containing 3
|
|
chr10_+_40225272
Show fit
|
1.24 |
ENSMUST00000044672.11
ENSMUST00000095743.4
|
Cdk19
|
cyclin-dependent kinase 19
|
|
chr6_+_136495784
Show fit
|
1.22 |
ENSMUST00000032335.13
ENSMUST00000185724.7
|
Atf7ip
|
activating transcription factor 7 interacting protein
|
|
chr7_-_3632826
Show fit
|
1.22 |
ENSMUST00000205596.2
ENSMUST00000155592.8
ENSMUST00000108641.10
|
Tfpt
|
TCF3 (E2A) fusion partner
|
|
chr18_-_34757653
Show fit
|
1.21 |
ENSMUST00000003876.10
ENSMUST00000115766.8
ENSMUST00000097626.10
ENSMUST00000115765.2
|
Brd8
|
bromodomain containing 8
|
|
chr14_+_25459630
Show fit
|
1.21 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1
|
|
chr14_-_30723292
Show fit
|
1.19 |
ENSMUST00000228736.2
ENSMUST00000226374.2
|
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae)
|
|
chr4_+_49632046
Show fit
|
1.18 |
ENSMUST00000156314.8
ENSMUST00000167496.8
ENSMUST00000029989.11
ENSMUST00000146547.2
|
Rnf20
|
ring finger protein 20
|
|
chr19_+_6107874
Show fit
|
1.17 |
ENSMUST00000178310.9
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived)
|
|
chr7_-_130964469
Show fit
|
1.17 |
ENSMUST00000059438.11
|
2310057M21Rik
|
RIKEN cDNA 2310057M21 gene
|
|
chr12_+_100165694
Show fit
|
1.16 |
ENSMUST00000110082.11
|
Calm1
|
calmodulin 1
|
|
chr12_+_51395047
Show fit
|
1.16 |
ENSMUST00000119211.8
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase
|
|
chr12_+_51395154
Show fit
|
1.15 |
ENSMUST00000121521.2
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase
|
|
chr5_+_34140777
Show fit
|
1.15 |
ENSMUST00000094869.12
ENSMUST00000114383.8
|
Gm1673
|
predicted gene 1673
|
|
chr19_-_29344694
Show fit
|
1.15 |
ENSMUST00000138051.2
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein
|
|
chrX_+_73314418
Show fit
|
1.14 |
ENSMUST00000008826.14
ENSMUST00000151702.8
ENSMUST00000074085.12
ENSMUST00000135690.2
|
Rpl10
|
ribosomal protein L10
|
|
chr17_+_6130061
Show fit
|
1.14 |
ENSMUST00000039487.10
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5
|
|
chr19_-_53378990
Show fit
|
1.12 |
ENSMUST00000025997.7
|
Smndc1
|
survival motor neuron domain containing 1
|
|
chr9_+_73020708
Show fit
|
1.12 |
ENSMUST00000169399.8
ENSMUST00000034738.14
|
Rsl24d1
|
ribosomal L24 domain containing 1
|
|
chr18_-_38471962
Show fit
|
1.10 |
ENSMUST00000139885.2
ENSMUST00000235590.2
ENSMUST00000237487.2
ENSMUST00000063814.15
|
Gnpda1
|
glucosamine-6-phosphate deaminase 1
|
|
chr15_-_77854711
Show fit
|
1.10 |
ENSMUST00000230419.2
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D
|
|
chr13_+_19132375
Show fit
|
1.10 |
ENSMUST00000239207.2
ENSMUST00000003345.10
ENSMUST00000200466.5
|
Amph
|
amphiphysin
|
|
chr9_+_57468217
Show fit
|
1.09 |
ENSMUST00000045791.11
ENSMUST00000216986.2
|
Scamp2
|
secretory carrier membrane protein 2
|
|
chr8_-_66939146
Show fit
|
1.03 |
ENSMUST00000026681.7
|
Tma16
|
translation machinery associated 16
|
|
chr9_-_36678868
Show fit
|
1.02 |
ENSMUST00000217599.2
ENSMUST00000120381.9
|
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr7_-_126574959
Show fit
|
1.02 |
ENSMUST00000206296.2
ENSMUST00000205437.3
|
Cdiptos
|
CDIP transferase, opposite strand
|
|
chr5_+_139186386
Show fit
|
1.01 |
ENSMUST00000058716.14
ENSMUST00000110883.9
ENSMUST00000110884.9
ENSMUST00000100517.10
ENSMUST00000127045.8
ENSMUST00000143562.8
ENSMUST00000078690.13
ENSMUST00000146715.8
|
Sun1
|
Sad1 and UNC84 domain containing 1
|
|
chr2_-_5849975
Show fit
|
1.00 |
ENSMUST00000043864.9
|
Cdc123
|
cell division cycle 123
|
|
chr15_-_79430942
Show fit
|
0.99 |
ENSMUST00000054014.9
ENSMUST00000229877.2
|
Ddx17
|
DEAD box helicase 17
|
|
chr7_-_28038129
Show fit
|
0.99 |
ENSMUST00000209141.2
ENSMUST00000003527.10
|
Supt5
|
suppressor of Ty 5, DSIF elongation factor subunit
|