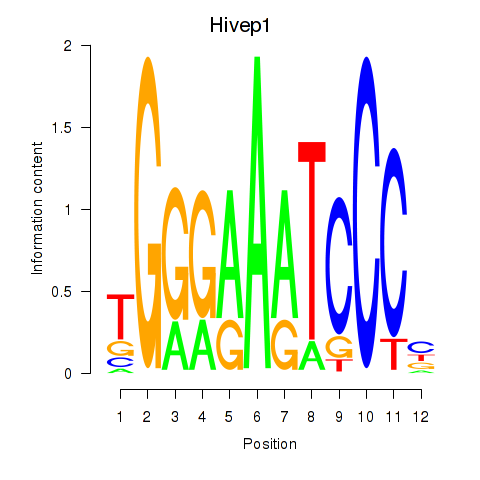
|
chr9_-_46146558
Show fit
|
3.17 |
ENSMUST00000121916.8
ENSMUST00000034586.9
|
Apoc3
|
apolipoprotein C-III
|
|
chr3_+_20039775
Show fit
|
2.87 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin
|
|
chr5_-_134776101
Show fit
|
2.84 |
ENSMUST00000015138.13
|
Eln
|
elastin
|
|
chr17_-_35081456
Show fit
|
2.81 |
ENSMUST00000025229.11
ENSMUST00000176203.9
ENSMUST00000128767.8
|
Cfb
|
complement factor B
|
|
chr10_-_95251327
Show fit
|
2.63 |
ENSMUST00000172070.8
ENSMUST00000150432.8
|
Socs2
|
suppressor of cytokine signaling 2
|
|
chr10_+_111342147
Show fit
|
2.42 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1
|
|
chr13_+_43938251
Show fit
|
2.38 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen
|
|
chr9_-_46146928
Show fit
|
2.28 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III
|
|
chr10_-_95251145
Show fit
|
2.18 |
ENSMUST00000119917.2
|
Socs2
|
suppressor of cytokine signaling 2
|
|
chr17_+_43978377
Show fit
|
2.17 |
ENSMUST00000233627.2
ENSMUST00000233437.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1
|
|
chr5_+_90708962
Show fit
|
2.11 |
ENSMUST00000094615.8
ENSMUST00000200765.2
|
Albfm1
|
albumin superfamily member 1
|
|
chr17_+_43978280
Show fit
|
2.07 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1
|
|
chr7_-_99276310
Show fit
|
1.99 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like
|
|
chr19_+_34268053
Show fit
|
1.61 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6)
|
|
chr4_-_57916283
Show fit
|
1.56 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene
|
|
chr11_+_78079562
Show fit
|
1.53 |
ENSMUST00000108322.9
|
Rab34
|
RAB34, member RAS oncogene family
|
|
chr19_+_34268071
Show fit
|
1.47 |
ENSMUST00000112472.4
ENSMUST00000235232.2
|
Fas
|
Fas (TNF receptor superfamily member 6)
|
|
chr9_-_70841881
Show fit
|
1.47 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic
|
|
chr16_-_30086317
Show fit
|
1.41 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2
|
|
chr17_+_35482063
Show fit
|
1.39 |
ENSMUST00000172503.3
|
H2-D1
|
histocompatibility 2, D region locus 1
|
|
chr17_+_35598583
Show fit
|
1.35 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4
|
|
chr1_+_192835414
Show fit
|
1.35 |
ENSMUST00000076521.7
|
Irf6
|
interferon regulatory factor 6
|
|
chr17_-_28736483
Show fit
|
1.35 |
ENSMUST00000114792.8
ENSMUST00000177939.8
|
Fkbp5
|
FK506 binding protein 5
|
|
chr6_+_71176811
Show fit
|
1.31 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver
|
|
chr17_+_35481702
Show fit
|
1.25 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1
|
|
chr14_+_55797934
Show fit
|
1.25 |
ENSMUST00000121622.8
ENSMUST00000143431.2
ENSMUST00000150481.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11
|
|
chr9_-_70842090
Show fit
|
1.24 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic
|
|
chr14_+_55798517
Show fit
|
1.24 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_21852508
Show fit
|
1.22 |
ENSMUST00000031678.10
|
Tspan12
|
tetraspanin 12
|
|
chr14_+_55798362
Show fit
|
1.22 |
ENSMUST00000072530.11
ENSMUST00000128490.9
|
Dcaf11
|
DDB1 and CUL4 associated factor 11
|
|
chr4_+_138181616
Show fit
|
1.15 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr6_-_21851827
Show fit
|
1.14 |
ENSMUST00000202353.2
ENSMUST00000134635.2
ENSMUST00000123116.8
ENSMUST00000120965.8
ENSMUST00000143531.2
|
Tspan12
|
tetraspanin 12
|
|
chr12_-_113561594
Show fit
|
1.12 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4
|
|
chr6_+_83034825
Show fit
|
1.11 |
ENSMUST00000077502.5
|
Dqx1
|
DEAQ RNA-dependent ATPase
|
|
chr11_+_78079631
Show fit
|
1.05 |
ENSMUST00000056241.12
ENSMUST00000207728.2
|
Rab34
|
RAB34, member RAS oncogene family
|
|
chr12_-_55539372
Show fit
|
0.97 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha
|
|
chr18_+_84106188
Show fit
|
0.93 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2
|
|
chrX_-_161747552
Show fit
|
0.92 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G
|
|
chr2_-_69172944
Show fit
|
0.92 |
ENSMUST00000102709.8
ENSMUST00000102710.10
ENSMUST00000180142.2
|
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr5_-_92496730
Show fit
|
0.91 |
ENSMUST00000038816.13
ENSMUST00000118006.3
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chr7_-_19363280
Show fit
|
0.90 |
ENSMUST00000094762.10
ENSMUST00000049912.15
ENSMUST00000098754.5
|
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B
|
|
chr4_-_133695264
Show fit
|
0.90 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2
|
|
chr11_+_78237492
Show fit
|
0.86 |
ENSMUST00000100755.4
|
Unc119
|
unc-119 lipid binding chaperone
|
|
chr3_-_132655954
Show fit
|
0.80 |
ENSMUST00000042744.16
ENSMUST00000117811.8
|
Npnt
|
nephronectin
|
|
chr17_-_34219225
Show fit
|
0.79 |
ENSMUST00000238098.2
ENSMUST00000087189.7
ENSMUST00000173075.3
ENSMUST00000172912.8
ENSMUST00000236740.2
ENSMUST00000025181.18
|
H2-K1
|
histocompatibility 2, K1, K region
|
|
chr11_+_101066867
Show fit
|
0.77 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1
|
|
chr19_+_38469504
Show fit
|
0.76 |
ENSMUST00000182481.8
|
Plce1
|
phospholipase C, epsilon 1
|
|
chr7_+_28466160
Show fit
|
0.70 |
ENSMUST00000122915.8
ENSMUST00000072965.5
ENSMUST00000170068.9
|
Sirt2
|
sirtuin 2
|
|
chr18_+_84106796
Show fit
|
0.69 |
ENSMUST00000235383.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2
|
|
chr7_+_19222972
Show fit
|
0.68 |
ENSMUST00000011407.8
|
Exoc3l2
|
exocyst complex component 3-like 2
|
|
chrX_+_35375751
Show fit
|
0.68 |
ENSMUST00000033418.8
|
Il13ra1
|
interleukin 13 receptor, alpha 1
|
|
chr17_+_35413415
Show fit
|
0.66 |
ENSMUST00000025262.6
ENSMUST00000173600.2
|
Ltb
|
lymphotoxin B
|
|
chr17_+_45866618
Show fit
|
0.66 |
ENSMUST00000024742.9
ENSMUST00000233929.2
|
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon
|
|
chr5_-_144294854
Show fit
|
0.66 |
ENSMUST00000055190.8
|
Baiap2l1
|
BAI1-associated protein 2-like 1
|
|
chr11_+_94102255
Show fit
|
0.66 |
ENSMUST00000041589.6
|
Tob1
|
transducer of ErbB-2.1
|
|
chr1_+_161796748
Show fit
|
0.65 |
ENSMUST00000111594.3
ENSMUST00000028021.7
ENSMUST00000193784.2
ENSMUST00000161826.2
|
Pigc
Gm38304
|
phosphatidylinositol glycan anchor biosynthesis, class C
predicted gene, 38304
|
|
chr12_-_113823290
Show fit
|
0.60 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17
|
|
chr12_-_113542610
Show fit
|
0.60 |
ENSMUST00000195468.6
ENSMUST00000103442.3
|
Ighv5-2
|
immunoglobulin heavy variable 5-2
|
|
chr1_+_161796854
Show fit
|
0.59 |
ENSMUST00000160881.2
ENSMUST00000159648.2
|
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C
|
|
chr11_+_70431063
Show fit
|
0.58 |
ENSMUST00000018429.12
ENSMUST00000108557.10
ENSMUST00000108556.2
|
Pld2
|
phospholipase D2
|
|
chr6_+_55313409
Show fit
|
0.58 |
ENSMUST00000004774.4
|
Aqp1
|
aquaporin 1
|
|
chr1_-_36312482
Show fit
|
0.57 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3
|
|
chr5_+_137348423
Show fit
|
0.56 |
ENSMUST00000111055.9
|
Ephb4
|
Eph receptor B4
|
|
chr2_-_5068807
Show fit
|
0.55 |
ENSMUST00000114996.8
|
Optn
|
optineurin
|
|
chr18_-_20192535
Show fit
|
0.54 |
ENSMUST00000075214.9
ENSMUST00000039247.11
|
Dsc2
|
desmocollin 2
|
|
chr5_-_144902598
Show fit
|
0.53 |
ENSMUST00000110677.8
ENSMUST00000085684.11
ENSMUST00000100461.7
|
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr5_-_116584765
Show fit
|
0.52 |
ENSMUST00000139425.2
|
Srrm4
|
serine/arginine repetitive matrix 4
|
|
chr5_+_137348363
Show fit
|
0.51 |
ENSMUST00000061244.15
|
Ephb4
|
Eph receptor B4
|
|
chr5_-_148489457
Show fit
|
0.50 |
ENSMUST00000079324.14
|
Ubl3
|
ubiquitin-like 3
|
|
chr4_-_82423985
Show fit
|
0.50 |
ENSMUST00000107245.9
ENSMUST00000107246.2
|
Nfib
|
nuclear factor I/B
|
|
chr17_+_35780977
Show fit
|
0.49 |
ENSMUST00000174525.8
ENSMUST00000068291.7
|
H2-Q10
|
histocompatibility 2, Q region locus 10
|
|
chr11_+_75546989
Show fit
|
0.48 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC
|
|
chr4_-_82423944
Show fit
|
0.47 |
ENSMUST00000107248.8
ENSMUST00000107247.8
|
Nfib
|
nuclear factor I/B
|
|
chr15_-_76501525
Show fit
|
0.46 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr11_+_75546671
Show fit
|
0.45 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC
|
|
chr10_-_27492827
Show fit
|
0.45 |
ENSMUST00000092639.12
|
Lama2
|
laminin, alpha 2
|
|
chr4_+_155646807
Show fit
|
0.45 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase
|
|
chr2_-_5068743
Show fit
|
0.45 |
ENSMUST00000027986.5
|
Optn
|
optineurin
|
|
chr9_+_5308828
Show fit
|
0.44 |
ENSMUST00000162846.8
ENSMUST00000027012.14
|
Casp4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr11_-_93846453
Show fit
|
0.43 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2
|
|
chr2_+_119803180
Show fit
|
0.43 |
ENSMUST00000066058.8
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1
|
|
chr3_+_122277372
Show fit
|
0.41 |
ENSMUST00000197073.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3
|
|
chr16_-_18052937
Show fit
|
0.41 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8
|
|
chr15_-_89033761
Show fit
|
0.40 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11
|
|
chrX_-_7242065
Show fit
|
0.40 |
ENSMUST00000191497.2
ENSMUST00000115744.2
|
Usp27x
|
ubiquitin specific peptidase 27, X chromosome
|
|
chr6_-_48817914
Show fit
|
0.38 |
ENSMUST00000164733.4
|
Tmem176b
|
transmembrane protein 176B
|
|
chr9_-_50679408
Show fit
|
0.38 |
ENSMUST00000177546.8
ENSMUST00000176238.2
|
1110032A03Rik
|
RIKEN cDNA 1110032A03 gene
|
|
chr17_+_56312672
Show fit
|
0.37 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing
|
|
chr6_-_48818006
Show fit
|
0.37 |
ENSMUST00000203229.3
|
Tmem176b
|
transmembrane protein 176B
|
|
chr18_+_37822865
Show fit
|
0.37 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2
|
|
chr9_-_44646487
Show fit
|
0.36 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1
|
|
chr11_-_70120503
Show fit
|
0.36 |
ENSMUST00000153449.2
ENSMUST00000000326.12
|
Bcl6b
|
B cell CLL/lymphoma 6, member B
|
|
chr19_-_5821442
Show fit
|
0.36 |
ENSMUST00000236978.2
|
Scyl1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr6_-_48818044
Show fit
|
0.35 |
ENSMUST00000101429.11
ENSMUST00000204073.3
|
Tmem176b
|
transmembrane protein 176B
|
|
chr12_-_113912416
Show fit
|
0.35 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1
|
|
chr8_+_95472218
Show fit
|
0.34 |
ENSMUST00000034231.4
|
Ccl22
|
chemokine (C-C motif) ligand 22
|
|
chr19_-_5135510
Show fit
|
0.33 |
ENSMUST00000140389.8
ENSMUST00000151413.2
ENSMUST00000077066.8
|
Tmem151a
|
transmembrane protein 151A
|
|
chr14_-_34225281
Show fit
|
0.33 |
ENSMUST00000171343.9
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A
|
|
chr7_-_90106375
Show fit
|
0.33 |
ENSMUST00000032844.7
|
Tmem126a
|
transmembrane protein 126A
|
|
chr2_-_118377500
Show fit
|
0.32 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor
|
|
chr4_-_40239778
Show fit
|
0.31 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58
|
|
chr19_-_5821373
Show fit
|
0.30 |
ENSMUST00000236297.2
ENSMUST00000236773.2
ENSMUST00000025890.10
|
Scyl1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr6_-_48818302
Show fit
|
0.30 |
ENSMUST00000203355.3
ENSMUST00000166247.8
|
Tmem176b
|
transmembrane protein 176B
|
|
chr16_-_28571820
Show fit
|
0.27 |
ENSMUST00000232352.2
|
Fgf12
|
fibroblast growth factor 12
|
|
chr19_+_6919229
Show fit
|
0.27 |
ENSMUST00000025910.13
|
Bad
|
BCL2-associated agonist of cell death
|
|
chr6_-_8778439
Show fit
|
0.26 |
ENSMUST00000115520.8
ENSMUST00000038403.12
ENSMUST00000115518.8
|
Ica1
|
islet cell autoantigen 1
|
|
chrX_-_23151771
Show fit
|
0.26 |
ENSMUST00000115319.9
|
Klhl13
|
kelch-like 13
|
|
chr2_+_32611067
Show fit
|
0.25 |
ENSMUST00000074248.11
|
Sh2d3c
|
SH2 domain containing 3C
|
|
chr11_+_5738480
Show fit
|
0.25 |
ENSMUST00000109845.8
ENSMUST00000020769.14
ENSMUST00000102928.5
|
Dbnl
|
drebrin-like
|
|
chr1_+_192984278
Show fit
|
0.24 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3
|
|
chr5_-_121975676
Show fit
|
0.24 |
ENSMUST00000197892.3
|
Sh2b3
|
SH2B adaptor protein 3
|
|
chr12_+_85871691
Show fit
|
0.24 |
ENSMUST00000040273.14
ENSMUST00000110224.8
ENSMUST00000142411.9
|
Ttll5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr12_+_111132779
Show fit
|
0.24 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3
|
|
chr11_-_5492175
Show fit
|
0.24 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117
|
|
chrX_-_8072714
Show fit
|
0.23 |
ENSMUST00000089403.10
ENSMUST00000077595.12
ENSMUST00000089402.10
ENSMUST00000082320.12
|
Porcn
|
porcupine O-acyltransferase
|
|
chr7_-_28465870
Show fit
|
0.22 |
ENSMUST00000085851.12
ENSMUST00000032815.11
|
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta
|
|
chr12_+_111132847
Show fit
|
0.20 |
ENSMUST00000021706.11
|
Traf3
|
TNF receptor-associated factor 3
|
|
chr3_+_96508400
Show fit
|
0.20 |
ENSMUST00000062058.5
|
Lix1l
|
Lix1-like
|
|
chr5_-_98178811
Show fit
|
0.20 |
ENSMUST00000031281.14
|
Antxr2
|
anthrax toxin receptor 2
|
|
chr14_+_52222283
Show fit
|
0.19 |
ENSMUST00000093813.12
ENSMUST00000100639.11
ENSMUST00000182909.8
ENSMUST00000182760.8
ENSMUST00000182061.8
ENSMUST00000182193.2
|
Arhgef40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr7_-_125799570
Show fit
|
0.18 |
ENSMUST00000009344.16
|
Xpo6
|
exportin 6
|
|
chr11_-_100302700
Show fit
|
0.18 |
ENSMUST00000141840.2
|
P3h4
|
prolyl 3-hydroxylase family member 4 (non-enzymatic)
|
|
chr15_-_38299934
Show fit
|
0.18 |
ENSMUST00000228772.2
|
Klf10
|
Kruppel-like factor 10
|
|
chr4_+_126156118
Show fit
|
0.17 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3
|
|
chr6_-_83513222
Show fit
|
0.16 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr7_-_79492091
Show fit
|
0.15 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase
|
|
chr19_+_6919691
Show fit
|
0.15 |
ENSMUST00000113426.8
ENSMUST00000238184.2
ENSMUST00000113423.10
ENSMUST00000237334.2
ENSMUST00000235240.2
|
Bad
|
BCL2-associated agonist of cell death
|
|
chr18_-_4352944
Show fit
|
0.15 |
ENSMUST00000025078.10
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr6_-_97464761
Show fit
|
0.15 |
ENSMUST00000032146.14
|
Frmd4b
|
FERM domain containing 4B
|
|
chr11_+_97697328
Show fit
|
0.15 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1
|
|
chr7_-_125799644
Show fit
|
0.15 |
ENSMUST00000168189.8
|
Xpo6
|
exportin 6
|
|
chr13_-_100382831
Show fit
|
0.14 |
ENSMUST00000049789.3
|
Naip5
|
NLR family, apoptosis inhibitory protein 5
|
|
chr19_-_6919755
Show fit
|
0.14 |
ENSMUST00000099782.10
|
Gpr137
|
G protein-coupled receptor 137
|
|
chr13_-_19521337
Show fit
|
0.14 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2
|
|
chr14_-_70666513
Show fit
|
0.12 |
ENSMUST00000226426.2
ENSMUST00000048129.6
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2
|
|
chr6_-_8778106
Show fit
|
0.12 |
ENSMUST00000151758.2
ENSMUST00000115519.8
ENSMUST00000153390.8
|
Ica1
|
islet cell autoantigen 1
|
|
chr11_+_98851238
Show fit
|
0.12 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha
|
|
chr5_-_98178834
Show fit
|
0.11 |
ENSMUST00000199088.2
|
Antxr2
|
anthrax toxin receptor 2
|
|
chr12_+_111132908
Show fit
|
0.11 |
ENSMUST00000139162.8
ENSMUST00000060274.7
|
Traf3
|
TNF receptor-associated factor 3
|
|
chr18_-_39062201
Show fit
|
0.10 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1
|
|
chr7_-_125799585
Show fit
|
0.10 |
ENSMUST00000163959.2
|
Xpo6
|
exportin 6
|
|
chr11_-_23720953
Show fit
|
0.09 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene
|
|
chr19_+_42040681
Show fit
|
0.08 |
ENSMUST00000164518.4
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene
|
|
chr7_+_27305222
Show fit
|
0.08 |
ENSMUST00000143499.8
ENSMUST00000108343.8
|
Akt2
|
thymoma viral proto-oncogene 2
|
|
chr15_+_76594810
Show fit
|
0.08 |
ENSMUST00000136840.8
ENSMUST00000127208.8
ENSMUST00000036423.15
ENSMUST00000137649.8
ENSMUST00000155225.2
ENSMUST00000155735.2
|
Lrrc14
|
leucine rich repeat containing 14
|
|
chr2_+_131792774
Show fit
|
0.08 |
ENSMUST00000110170.8
ENSMUST00000110172.9
ENSMUST00000110171.9
|
Prnd
|
prion like protein doppel
|
|
chr19_+_24853039
Show fit
|
0.07 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053
|
|
chr1_-_172418058
Show fit
|
0.07 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8
|
|
chr2_-_103133524
Show fit
|
0.07 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor
|
|
chr12_+_36364136
Show fit
|
0.07 |
ENSMUST00000041407.7
|
Sostdc1
|
sclerostin domain containing 1
|
|
chr10_-_27492792
Show fit
|
0.07 |
ENSMUST00000189575.2
|
Lama2
|
laminin, alpha 2
|
|
chr7_+_27305271
Show fit
|
0.06 |
ENSMUST00000051356.12
|
Akt2
|
thymoma viral proto-oncogene 2
|
|
chrX_+_82898974
Show fit
|
0.06 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy
|
|
chr15_+_102379621
Show fit
|
0.06 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2
|
|
chr10_+_119828821
Show fit
|
0.05 |
ENSMUST00000105261.9
|
Grip1
|
glutamate receptor interacting protein 1
|
|
chr15_-_79048674
Show fit
|
0.05 |
ENSMUST00000230261.2
ENSMUST00000040019.5
|
Sox10
|
SRY (sex determining region Y)-box 10
|
|
chr15_+_102379503
Show fit
|
0.05 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2
|
|
chr11_-_99176086
Show fit
|
0.05 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24
|
|
chr15_-_78090591
Show fit
|
0.04 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin
|
|
chr2_-_119377675
Show fit
|
0.03 |
ENSMUST00000133668.2
|
Exd1
|
exonuclease 3'-5' domain containing 1
|
|
chr10_+_127216459
Show fit
|
0.03 |
ENSMUST00000166820.8
|
R3hdm2
|
R3H domain containing 2
|
|
chr5_+_137059127
Show fit
|
0.03 |
ENSMUST00000041543.9
ENSMUST00000186451.2
|
Vgf
|
VGF nerve growth factor inducible
|
|
chr4_+_128777339
Show fit
|
0.03 |
ENSMUST00000035667.9
|
Trim62
|
tripartite motif-containing 62
|
|
chr4_-_40239700
Show fit
|
0.02 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58
|
|
chr2_-_103133503
Show fit
|
0.02 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor
|
|
chr3_+_88857929
Show fit
|
0.01 |
ENSMUST00000186583.7
|
Ash1l
|
ASH1 like histone lysine methyltransferase
|
|
chr12_+_98886826
Show fit
|
0.01 |
ENSMUST00000085109.10
ENSMUST00000079146.13
|
Ttc8
|
tetratricopeptide repeat domain 8
|
|
chr10_-_81463631
Show fit
|
0.01 |
ENSMUST00000042923.9
|
Sirt6
|
sirtuin 6
|
|
chr10_-_81463091
Show fit
|
0.01 |
ENSMUST00000143424.2
ENSMUST00000119324.8
|
Sirt6
|
sirtuin 6
|
|
chr10_+_119828867
Show fit
|
0.01 |
ENSMUST00000154238.8
|
Grip1
|
glutamate receptor interacting protein 1
|
|
chr7_-_30371021
Show fit
|
0.01 |
ENSMUST00000051495.7
|
Pmis2
|
PMIS2 transmembrane protein
|
|
chr4_-_120672900
Show fit
|
0.01 |
ENSMUST00000120779.8
|
Nfyc
|
nuclear transcription factor-Y gamma
|
|
chr2_-_57004933
Show fit
|
0.01 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr1_+_186947934
Show fit
|
0.00 |
ENSMUST00000160471.8
|
Gpatch2
|
G patch domain containing 2
|
|
chr18_-_34784746
Show fit
|
0.00 |
ENSMUST00000025228.12
ENSMUST00000133181.2
|
Cdc23
|
CDC23 cell division cycle 23
|