Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hmx2
Z-value: 2.21
Transcription factors associated with Hmx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx2
|
ENSMUSG00000050100.15 | Hmx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx2 | mm39_v1_chr7_+_131150484_131150502 | -0.29 | 9.2e-02 | Click! |
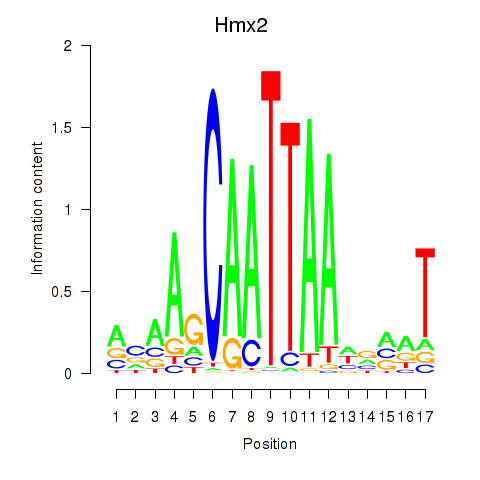
Activity profile of Hmx2 motif
Sorted Z-values of Hmx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_146016064 | 18.93 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr4_-_61972348 | 11.14 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr14_+_51333816 | 10.48 |
ENSMUST00000169895.3
|
Rnase4
|
ribonuclease, RNase A family 4 |
| chr13_-_24098981 | 10.13 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr13_-_24098951 | 9.51 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr19_-_8382424 | 7.99 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr19_-_7943365 | 7.35 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr19_+_12610870 | 7.24 |
ENSMUST00000119960.2
|
Glyat
|
glycine-N-acyltransferase |
| chr19_+_12610668 | 6.53 |
ENSMUST00000044976.12
|
Glyat
|
glycine-N-acyltransferase |
| chr19_-_58442866 | 5.44 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr2_-_23938869 | 5.10 |
ENSMUST00000114497.2
|
Hnmt
|
histamine N-methyltransferase |
| chr5_+_9163244 | 4.81 |
ENSMUST00000198935.2
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr2_-_23939401 | 4.71 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr7_+_119206233 | 4.45 |
ENSMUST00000126367.8
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr16_-_56987670 | 4.38 |
ENSMUST00000023432.10
|
Nit2
|
nitrilase family, member 2 |
| chr10_-_128425519 | 4.31 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr3_+_94280101 | 4.28 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr4_-_96441854 | 4.24 |
ENSMUST00000030303.6
|
Cyp2j6
|
cytochrome P450, family 2, subfamily j, polypeptide 6 |
| chr1_-_162694076 | 4.13 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr13_-_63036096 | 3.73 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr1_+_91226076 | 3.20 |
ENSMUST00000142488.8
ENSMUST00000124832.8 ENSMUST00000147523.8 |
Scly
|
selenocysteine lyase |
| chr2_+_167263626 | 3.17 |
ENSMUST00000047815.13
ENSMUST00000109218.7 ENSMUST00000073873.5 |
Slc9a8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8 |
| chr1_-_172460497 | 2.96 |
ENSMUST00000027826.7
|
Dusp23
|
dual specificity phosphatase 23 |
| chr3_+_20011405 | 2.95 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr10_-_19783391 | 2.91 |
ENSMUST00000166511.9
ENSMUST00000020182.16 |
Pex7
|
peroxisomal biogenesis factor 7 |
| chr1_+_91226058 | 2.87 |
ENSMUST00000027532.13
|
Scly
|
selenocysteine lyase |
| chrX_+_138464065 | 2.75 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr11_-_106503754 | 2.71 |
ENSMUST00000042780.14
|
Tex2
|
testis expressed gene 2 |
| chr7_-_44711771 | 2.40 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr2_-_127286444 | 2.23 |
ENSMUST00000028848.4
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr8_+_25507227 | 2.20 |
ENSMUST00000033961.7
ENSMUST00000210536.2 |
Tm2d2
|
TM2 domain containing 2 |
| chr2_-_75768752 | 2.15 |
ENSMUST00000099996.5
|
Ttc30b
|
tetratricopeptide repeat domain 30B |
| chr11_-_106504196 | 2.12 |
ENSMUST00000128933.2
|
Tex2
|
testis expressed gene 2 |
| chr3_+_20011201 | 2.05 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr15_-_74869684 | 1.98 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr6_-_108162513 | 1.88 |
ENSMUST00000167338.8
ENSMUST00000172188.2 ENSMUST00000032191.16 |
Sumf1
|
sulfatase modifying factor 1 |
| chr5_-_87739442 | 1.82 |
ENSMUST00000031201.9
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr17_-_34847354 | 1.78 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr11_-_46280336 | 1.76 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr19_-_44095840 | 1.75 |
ENSMUST00000119591.2
ENSMUST00000026217.11 |
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr16_+_34815177 | 1.74 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chr2_-_26823793 | 1.71 |
ENSMUST00000154651.2
ENSMUST00000015011.10 |
Surf4
|
surfeit gene 4 |
| chr5_+_8710059 | 1.68 |
ENSMUST00000047753.5
|
Abcb1a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1A |
| chr4_+_145595364 | 1.68 |
ENSMUST00000123460.2
|
Zfp986
|
zinc finger protein 986 |
| chr4_+_145241454 | 1.64 |
ENSMUST00000105741.2
|
Zfp990
|
zinc finger protein 990 |
| chrX_-_162810959 | 1.64 |
ENSMUST00000033739.5
|
Car5b
|
carbonic anhydrase 5b, mitochondrial |
| chr9_-_15212745 | 1.55 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr11_-_46280298 | 1.47 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr10_+_29019645 | 1.45 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chrX_-_55643429 | 1.44 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr3_+_106628987 | 1.44 |
ENSMUST00000130105.2
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr1_+_107289659 | 1.42 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr10_+_121200984 | 1.38 |
ENSMUST00000040344.7
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr12_-_98867737 | 1.37 |
ENSMUST00000223282.2
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chrM_+_7758 | 1.32 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr11_-_46280281 | 1.29 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr6_+_40619913 | 1.28 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chrX_+_37405054 | 1.28 |
ENSMUST00000016471.9
ENSMUST00000115134.2 |
Atp1b4
|
ATPase, (Na+)/K+ transporting, beta 4 polypeptide |
| chr11_-_114939152 | 1.26 |
ENSMUST00000092459.4
|
Cd300ld3
|
CD300 molecule like family member D3 |
| chr12_+_84498196 | 1.24 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr14_+_48683581 | 1.22 |
ENSMUST00000227440.2
ENSMUST00000124720.8 ENSMUST00000226422.2 ENSMUST00000226400.2 |
Tmem260
|
transmembrane protein 260 |
| chrM_+_7779 | 1.22 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr4_+_145237329 | 1.21 |
ENSMUST00000105742.8
ENSMUST00000136309.8 |
Zfp990
|
zinc finger protein 990 |
| chr2_+_30282266 | 1.15 |
ENSMUST00000028209.15
|
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr2_-_111324108 | 1.13 |
ENSMUST00000208881.2
ENSMUST00000208695.2 ENSMUST00000217611.2 |
Olfr1290
|
olfactory receptor 1290 |
| chr4_-_128856213 | 1.11 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr13_+_23868175 | 1.10 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr18_+_63110899 | 1.09 |
ENSMUST00000025474.14
|
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr11_-_100713348 | 1.08 |
ENSMUST00000107358.9
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr19_+_29923182 | 1.06 |
ENSMUST00000025724.9
|
Il33
|
interleukin 33 |
| chr8_-_23184070 | 1.05 |
ENSMUST00000131767.2
|
Ikbkb
|
inhibitor of kappaB kinase beta |
| chr2_+_30282414 | 1.02 |
ENSMUST00000123202.8
ENSMUST00000113612.10 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr11_+_58062467 | 1.02 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr5_-_28415020 | 1.01 |
ENSMUST00000118882.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr5_-_28415166 | 0.97 |
ENSMUST00000117098.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr12_+_72488625 | 0.94 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr4_+_146033882 | 0.94 |
ENSMUST00000105730.2
ENSMUST00000091878.6 |
Zfp987
|
zinc finger protein 987 |
| chr4_+_122730027 | 0.92 |
ENSMUST00000030412.11
ENSMUST00000121870.8 ENSMUST00000097902.5 |
Ppt1
|
palmitoyl-protein thioesterase 1 |
| chr17_+_14087827 | 0.91 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr18_+_63111005 | 0.89 |
ENSMUST00000235372.2
ENSMUST00000237483.2 |
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr4_-_42874178 | 0.88 |
ENSMUST00000107978.2
|
Fam205c
|
family with sequence similarity 205, member C |
| chr4_+_147576874 | 0.86 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr4_-_42874197 | 0.85 |
ENSMUST00000055944.11
|
Fam205c
|
family with sequence similarity 205, member C |
| chr18_-_63520195 | 0.84 |
ENSMUST00000047480.13
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr14_-_52728503 | 0.83 |
ENSMUST00000073571.6
|
Olfr1507
|
olfactory receptor 1507 |
| chr18_-_88912415 | 0.83 |
ENSMUST00000145120.3
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr16_-_44153288 | 0.83 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr17_+_36152559 | 0.82 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr18_+_37952556 | 0.81 |
ENSMUST00000055935.11
|
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr11_-_77971186 | 0.81 |
ENSMUST00000021183.4
|
Eral1
|
Era (G-protein)-like 1 (E. coli) |
| chr3_-_65300000 | 0.78 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr5_+_29400981 | 0.78 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chrX_+_149377416 | 0.78 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr4_-_146993984 | 0.76 |
ENSMUST00000238583.2
ENSMUST00000049821.4 |
Gm21411
|
predicted gene, 21411 |
| chr6_+_134617903 | 0.70 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr7_-_106531426 | 0.70 |
ENSMUST00000215468.2
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr18_+_63110924 | 0.67 |
ENSMUST00000150267.2
ENSMUST00000236925.2 |
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr6_-_119940694 | 0.67 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr11_-_114928545 | 0.67 |
ENSMUST00000100239.3
|
Cd300ld5
|
CD300 molecule like family member D5 |
| chr2_-_32977182 | 0.66 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr7_-_78432774 | 0.66 |
ENSMUST00000032841.7
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr18_+_37952596 | 0.65 |
ENSMUST00000193890.2
ENSMUST00000193941.2 |
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr8_+_72676649 | 0.64 |
ENSMUST00000119003.2
|
Zfp617
|
zinc finger protein 617 |
| chr9_-_75518387 | 0.63 |
ENSMUST00000215462.2
ENSMUST00000217233.2 ENSMUST00000215614.2 |
Tmod2
|
tropomodulin 2 |
| chr9_-_75518585 | 0.62 |
ENSMUST00000098552.10
ENSMUST00000064433.11 |
Tmod2
|
tropomodulin 2 |
| chr16_+_32315090 | 0.62 |
ENSMUST00000231510.2
|
Zdhhc19
|
zinc finger, DHHC domain containing 19 |
| chr16_-_44153498 | 0.61 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr4_+_33081505 | 0.61 |
ENSMUST00000147889.2
|
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr11_+_115197980 | 0.61 |
ENSMUST00000055490.9
|
Otop2
|
otopetrin 2 |
| chr4_+_147106307 | 0.61 |
ENSMUST00000075775.6
|
Rex2
|
reduced expression 2 |
| chr4_+_146093394 | 0.61 |
ENSMUST00000168483.9
|
Zfp600
|
zinc finger protein 600 |
| chr1_+_46105898 | 0.60 |
ENSMUST00000069293.10
|
Dnah7b
|
dynein, axonemal, heavy chain 7B |
| chr14_-_52011035 | 0.60 |
ENSMUST00000073860.6
|
Ang4
|
angiogenin, ribonuclease A family, member 4 |
| chr1_+_46106006 | 0.59 |
ENSMUST00000238212.2
|
Dnah7b
|
dynein, axonemal, heavy chain 7B |
| chr18_-_66072119 | 0.58 |
ENSMUST00000025396.5
|
Rax
|
retina and anterior neural fold homeobox |
| chr6_+_40448286 | 0.58 |
ENSMUST00000114779.9
|
Ssbp1
|
single-stranded DNA binding protein 1 |
| chr3_+_66893280 | 0.58 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr9_-_75518357 | 0.57 |
ENSMUST00000213565.2
|
Tmod2
|
tropomodulin 2 |
| chr16_+_17437367 | 0.56 |
ENSMUST00000231887.2
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr9_+_19047613 | 0.56 |
ENSMUST00000215699.2
|
Olfr837
|
olfactory receptor 837 |
| chr8_+_91555449 | 0.56 |
ENSMUST00000109614.9
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_31117617 | 0.55 |
ENSMUST00000202567.2
ENSMUST00000074840.12 |
Preb
|
prolactin regulatory element binding |
| chr2_+_26824040 | 0.54 |
ENSMUST00000153771.8
ENSMUST00000055406.9 |
Stkld1
|
serine/threonine kinase-like domain containing 1 |
| chr9_-_54408780 | 0.54 |
ENSMUST00000118600.8
ENSMUST00000118163.8 |
Dmxl2
|
Dmx-like 2 |
| chr8_-_55177510 | 0.53 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr2_-_17465410 | 0.53 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr13_-_76453191 | 0.52 |
ENSMUST00000208418.2
|
Fam81b
|
family with sequence similarity 81, member B |
| chr11_-_99241924 | 0.51 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
| chr8_+_26298502 | 0.50 |
ENSMUST00000033979.6
|
Star
|
steroidogenic acute regulatory protein |
| chr17_-_20701593 | 0.50 |
ENSMUST00000167314.3
ENSMUST00000232850.2 ENSMUST00000233382.2 ENSMUST00000233572.2 ENSMUST00000233830.2 |
Vmn2r108
|
vomeronasal 2, receptor 108 |
| chr3_+_18108313 | 0.48 |
ENSMUST00000026120.8
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr18_-_34712123 | 0.48 |
ENSMUST00000079287.12
|
Nme5
|
NME/NM23 family member 5 |
| chr10_+_69761630 | 0.47 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr4_+_90107057 | 0.47 |
ENSMUST00000107129.2
|
Zfp352
|
zinc finger protein 352 |
| chr7_+_102965522 | 0.46 |
ENSMUST00000216456.2
|
Olfr597
|
olfactory receptor 597 |
| chr9_+_51958453 | 0.46 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr7_-_10803618 | 0.46 |
ENSMUST00000165848.10
|
Zscan4-ps1
|
zinc finger and SCAN domain containing 4, pseudogene 1 |
| chr4_-_15149755 | 0.46 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr14_+_48683797 | 0.46 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chr6_+_58239022 | 0.46 |
ENSMUST00000227805.2
|
Vmn1r28
|
vomeronasal 1 receptor 28 |
| chr5_-_110242267 | 0.46 |
ENSMUST00000211397.2
ENSMUST00000210052.2 |
Gm35315
|
predicted gene, 35315 |
| chr11_+_107370375 | 0.45 |
ENSMUST00000106750.5
|
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr2_+_177409837 | 0.45 |
ENSMUST00000108942.5
|
Gm14322
|
predicted gene 14322 |
| chr5_+_114518653 | 0.44 |
ENSMUST00000074002.12
|
Ube3b
|
ubiquitin protein ligase E3B |
| chr9_+_92339422 | 0.44 |
ENSMUST00000034941.9
|
Plscr4
|
phospholipid scramblase 4 |
| chr4_+_147445744 | 0.43 |
ENSMUST00000133078.8
ENSMUST00000154154.2 |
Zfp978
|
zinc finger protein 978 |
| chr2_+_57887896 | 0.42 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr12_-_40249314 | 0.42 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr9_+_19533591 | 0.42 |
ENSMUST00000215372.2
|
Zfp317
|
zinc finger protein 317 |
| chr1_-_58009263 | 0.42 |
ENSMUST00000114410.10
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr4_-_43840201 | 0.41 |
ENSMUST00000214281.2
|
Olfr157
|
olfactory receptor 157 |
| chr14_+_52964069 | 0.40 |
ENSMUST00000184883.2
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr18_-_66155651 | 0.40 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr18_-_66072168 | 0.40 |
ENSMUST00000238035.2
|
Rax
|
retina and anterior neural fold homeobox |
| chr2_-_32976378 | 0.39 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr4_-_147787010 | 0.38 |
ENSMUST00000117638.2
|
Zfp534
|
zinc finger protein 534 |
| chr17_-_37613523 | 0.38 |
ENSMUST00000215392.2
|
Olfr101
|
olfactory receptor 101 |
| chr2_-_88909981 | 0.37 |
ENSMUST00000213724.2
|
Olfr1219
|
olfactory receptor 1219 |
| chr6_+_40448334 | 0.37 |
ENSMUST00000031971.13
|
Ssbp1
|
single-stranded DNA binding protein 1 |
| chr2_+_85835884 | 0.37 |
ENSMUST00000111589.3
|
Olfr1032
|
olfactory receptor 1032 |
| chr7_-_12537533 | 0.37 |
ENSMUST00000210650.2
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr11_-_114917838 | 0.36 |
ENSMUST00000100240.3
|
Cd300ld4
|
CD300 molecule like family member D4 |
| chr6_-_113478779 | 0.36 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr7_+_23752679 | 0.36 |
ENSMUST00000236959.2
|
Vmn1r183
|
vomeronasal 1 receptor 183 |
| chr6_-_69920632 | 0.36 |
ENSMUST00000198880.5
ENSMUST00000103371.3 |
Igkv12-38
|
immunoglobulin kappa chain variable 12-38 |
| chr16_+_32315076 | 0.36 |
ENSMUST00000064192.8
|
Zdhhc19
|
zinc finger, DHHC domain containing 19 |
| chr11_-_67661945 | 0.35 |
ENSMUST00000051765.9
|
Glp2r
|
glucagon-like peptide 2 receptor |
| chr5_-_28672091 | 0.35 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr7_+_102834010 | 0.35 |
ENSMUST00000106893.3
|
Olfr592
|
olfactory receptor 592 |
| chr7_+_139913930 | 0.33 |
ENSMUST00000214858.2
|
Olfr527
|
olfactory receptor 527 |
| chr5_+_102916637 | 0.32 |
ENSMUST00000112852.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr7_-_103191924 | 0.32 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612 |
| chr4_+_146599377 | 0.31 |
ENSMUST00000140089.7
ENSMUST00000179175.3 |
Zfp981
|
zinc finger protein 981 |
| chr10_-_127147609 | 0.31 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chr6_+_58239367 | 0.30 |
ENSMUST00000226813.2
|
Vmn1r28
|
vomeronasal 1 receptor 28 |
| chr1_+_164103127 | 0.30 |
ENSMUST00000027867.7
|
Ccdc181
|
coiled-coil domain containing 181 |
| chr6_+_129022843 | 0.29 |
ENSMUST00000032257.10
ENSMUST00000204320.2 |
Klrb1f
|
killer cell lectin-like receptor subfamily B member 1F |
| chr14_+_53666165 | 0.28 |
ENSMUST00000180549.3
|
Trav6-3
|
T cell receptor alpha variable 6-3 |
| chr17_+_46739852 | 0.27 |
ENSMUST00000113465.10
ENSMUST00000024764.12 ENSMUST00000165993.2 |
Crip3
|
cysteine-rich protein 3 |
| chr1_+_177557380 | 0.27 |
ENSMUST00000016106.6
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr14_-_52616625 | 0.26 |
ENSMUST00000214980.2
|
Olfr1512
|
olfactory receptor 1512 |
| chr14_+_51366306 | 0.26 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr4_+_147390131 | 0.26 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988 |
| chr7_-_24245419 | 0.25 |
ENSMUST00000011776.8
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr7_+_11464193 | 0.24 |
ENSMUST00000226516.2
|
Vmn1r73
|
vomeronasal 1 receptor 73 |
| chr7_-_12103200 | 0.24 |
ENSMUST00000228653.2
ENSMUST00000227427.2 ENSMUST00000226408.2 |
Vmn1r84
|
vomeronasal 1 receptor 84 |
| chrX_+_105059305 | 0.23 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr17_+_30940015 | 0.23 |
ENSMUST00000236948.2
|
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr15_-_34356567 | 0.23 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr3_-_64322789 | 0.22 |
ENSMUST00000175724.9
|
Vmn2r4
|
vomeronasal 2, receptor 4 |
| chr4_+_138606671 | 0.22 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr4_+_147056433 | 0.22 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989 |
| chr16_-_19060440 | 0.22 |
ENSMUST00000103751.3
|
Iglv3
|
immunoglobulin lambda variable 3 |
| chr15_-_12493574 | 0.21 |
ENSMUST00000186113.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr10_+_69761597 | 0.21 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr6_-_129362460 | 0.20 |
ENSMUST00000032261.9
|
Clec12b
|
C-type lectin domain family 12, member B |
| chrX_+_11184495 | 0.20 |
ENSMUST00000179859.2
|
H2al1g
|
H2A histone family member L1G |
| chr13_+_23191826 | 0.20 |
ENSMUST00000228758.2
ENSMUST00000228031.2 ENSMUST00000227573.2 |
Vmn1r213
|
vomeronasal 1 receptor 213 |
| chr2_-_73283010 | 0.19 |
ENSMUST00000151939.2
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr4_+_145397238 | 0.18 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 9.8 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 1.9 | 11.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 1.0 | 15.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.9 | 3.7 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.9 | 6.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.8 | 4.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.6 | 1.8 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.5 | 4.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.4 | 1.7 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.4 | 4.3 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.4 | 2.9 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 1.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.3 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.3 | 1.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 4.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 4.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 0.4 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.2 | 1.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 1.8 | GO:0072641 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.2 | 1.1 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.2 | 0.9 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 3.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 5.0 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.7 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 1.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 19.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.0 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 1.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.5 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 2.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 14.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 2.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.5 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 10.5 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.1 | 0.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 4.4 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 2.1 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.0 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 3.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 1.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.8 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.7 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 2.5 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 2.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 2.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 5.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 5.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 7.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 2.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 6.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 4.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.4 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 18.9 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 3.4 | 13.8 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 2.2 | 11.1 | GO:0005186 | pheromone activity(GO:0005186) |
| 2.0 | 6.1 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 1.2 | 19.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.1 | 4.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.0 | 15.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 4.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 1.7 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.6 | 4.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.5 | 2.2 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.5 | 2.6 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.5 | 4.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.5 | 2.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.5 | 5.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 4.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 3.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 2.9 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 4.5 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.3 | 1.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 2.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 1.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 1.3 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.2 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.4 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 1.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 9.8 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 0.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 9.4 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 0.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 3.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 3.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 4.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 2.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 5.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 2.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 1.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 14.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 4.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 5.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 4.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 5.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.1 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 4.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 3.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |