Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
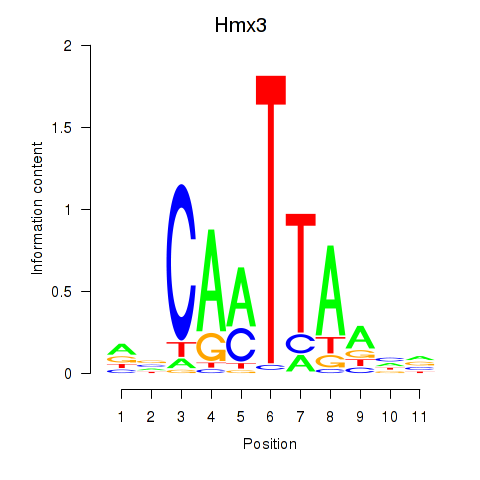
Results for Hmx3
Z-value: 0.61
Transcription factors associated with Hmx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx3
|
ENSMUSG00000040148.6 | Hmx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx3 | mm39_v1_chr7_+_131144596_131144596 | 0.29 | 8.4e-02 | Click! |
Activity profile of Hmx3 motif
Sorted Z-values of Hmx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_150510116 | 1.69 |
ENSMUST00000028944.4
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr1_-_45542442 | 1.45 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr11_-_20282684 | 1.43 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr6_+_86348286 | 1.37 |
ENSMUST00000089558.7
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr7_+_79944198 | 1.24 |
ENSMUST00000163812.9
ENSMUST00000047558.14 ENSMUST00000174199.8 ENSMUST00000173824.8 ENSMUST00000174172.8 |
Prc1
|
protein regulator of cytokinesis 1 |
| chr7_+_126808016 | 0.90 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr11_+_114559350 | 0.87 |
ENSMUST00000106602.10
ENSMUST00000077915.10 ENSMUST00000106599.8 ENSMUST00000082092.5 |
Rpl38
|
ribosomal protein L38 |
| chr5_+_146885450 | 0.87 |
ENSMUST00000146511.8
ENSMUST00000132102.2 |
Gtf3a
|
general transcription factor III A |
| chr5_+_110987839 | 0.85 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr7_+_25380195 | 0.73 |
ENSMUST00000205658.2
|
B9d2
|
B9 protein domain 2 |
| chr1_-_93088562 | 0.64 |
ENSMUST00000143419.2
|
Mab21l4
|
mab-21-like 4 |
| chr7_-_126807581 | 0.64 |
ENSMUST00000120705.3
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr7_+_78922947 | 0.61 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr7_+_24967094 | 0.59 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor |
| chr7_-_131918926 | 0.58 |
ENSMUST00000080215.6
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chr3_+_96537235 | 0.57 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr15_+_9436114 | 0.56 |
ENSMUST00000042360.5
ENSMUST00000226688.2 |
Capsl
|
calcyphosine-like |
| chr8_-_23727639 | 0.56 |
ENSMUST00000033950.7
|
Gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr7_+_25380263 | 0.55 |
ENSMUST00000205325.2
ENSMUST00000206913.2 |
B9d2
|
B9 protein domain 2 |
| chr2_+_3115250 | 0.54 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr3_+_96537484 | 0.54 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr9_-_56151334 | 0.52 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr6_-_87312743 | 0.49 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr8_+_107757847 | 0.45 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr9_-_110453427 | 0.44 |
ENSMUST00000196876.2
ENSMUST00000035069.14 |
Nradd
|
neurotrophin receptor associated death domain |
| chr1_-_149836974 | 0.43 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr7_-_30259025 | 0.41 |
ENSMUST00000043975.11
ENSMUST00000156241.2 |
Lin37
|
lin-37 homolog (C. elegans) |
| chrX_+_149377416 | 0.41 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr5_-_53864874 | 0.40 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chrX_+_158491589 | 0.39 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_-_30259253 | 0.39 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr15_-_79658608 | 0.37 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr4_-_62389098 | 0.35 |
ENSMUST00000135811.2
ENSMUST00000120095.8 ENSMUST00000030087.14 ENSMUST00000107452.8 ENSMUST00000155522.8 |
Wdr31
|
WD repeat domain 31 |
| chr11_+_96209093 | 0.34 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr19_+_13208692 | 0.34 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr2_+_130266253 | 0.33 |
ENSMUST00000128994.8
ENSMUST00000028900.11 |
Vps16
|
VSP16 CORVET/HOPS core subunit |
| chr6_-_129599645 | 0.33 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr10_+_99099084 | 0.32 |
ENSMUST00000020118.5
ENSMUST00000220291.2 |
Dusp6
|
dual specificity phosphatase 6 |
| chr12_+_108145802 | 0.32 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr4_+_80752360 | 0.29 |
ENSMUST00000133655.8
ENSMUST00000006151.13 |
Tyrp1
|
tyrosinase-related protein 1 |
| chr4_+_19818718 | 0.29 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr15_-_79658584 | 0.28 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr19_+_8779903 | 0.28 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr12_+_108145997 | 0.27 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr9_+_106048116 | 0.26 |
ENSMUST00000020490.13
|
Wdr82
|
WD repeat domain containing 82 |
| chr19_+_41921903 | 0.25 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chr14_+_54598283 | 0.25 |
ENSMUST00000000985.7
|
Oxa1l
|
oxidase assembly 1-like |
| chr4_+_80752535 | 0.24 |
ENSMUST00000102831.8
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr2_-_152857239 | 0.24 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr10_-_80257681 | 0.22 |
ENSMUST00000156244.2
|
Tcf3
|
transcription factor 3 |
| chr11_+_6366259 | 0.21 |
ENSMUST00000213200.2
|
Ppia
|
peptidylprolyl isomerase A |
| chr9_-_123507847 | 0.20 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr2_+_74528071 | 0.20 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9 |
| chr3_-_49711765 | 0.20 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr7_+_3648264 | 0.20 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr15_+_21111428 | 0.19 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr3_-_154760978 | 0.17 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr3_-_49711706 | 0.16 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr10_+_115405891 | 0.16 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr7_+_83281193 | 0.16 |
ENSMUST00000117410.2
|
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr13_+_23930717 | 0.15 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr19_-_13126896 | 0.15 |
ENSMUST00000213493.2
|
Olfr1459
|
olfactory receptor 1459 |
| chr5_-_87630117 | 0.13 |
ENSMUST00000079811.13
ENSMUST00000144144.3 |
Ugt2a2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr2_+_59442378 | 0.13 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr11_-_99265721 | 0.12 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr8_-_33374825 | 0.09 |
ENSMUST00000238791.2
|
Nrg1
|
neuregulin 1 |
| chr17_-_36008863 | 0.08 |
ENSMUST00000146472.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr2_-_45002902 | 0.08 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr15_+_66542598 | 0.08 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin |
| chr12_+_52746158 | 0.06 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr4_-_45532470 | 0.06 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr9_+_21867043 | 0.05 |
ENSMUST00000053583.7
|
Swsap1
|
SWIM type zinc finger 7 associated protein 1 |
| chr4_+_65523223 | 0.04 |
ENSMUST00000050850.14
ENSMUST00000107366.2 |
Trim32
|
tripartite motif-containing 32 |
| chr8_+_10299288 | 0.01 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.5 | 1.4 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.4 | 1.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 0.8 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.2 | 0.5 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.3 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.1 | 0.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.5 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.9 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 0.6 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.4 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.4 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.6 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.1 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 0.9 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.2 | 0.6 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 0.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.3 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.1 | 1.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.5 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |